Solutions to additional exercises
Genome Evolution
Additional Exercise: WGDs in Populus trichocarpa
- The same basic steps are necessary to perform this analysis as in the tutorial itself
- Though the interpretation of the results is slightly more difficult
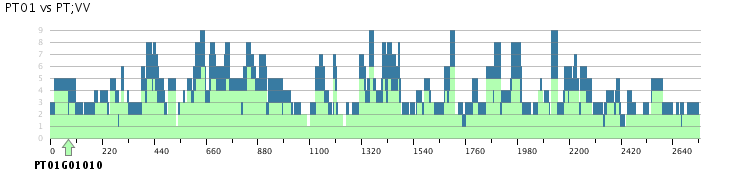
- Interpretation of the skyline plot
- Contrary to the skyline plot in the tutorial poplar-grapevine plots reveal several times 2:1, 3-4:2, 5-6:3 ratios of segments. This already is a strong indication that there was a genome doubling.
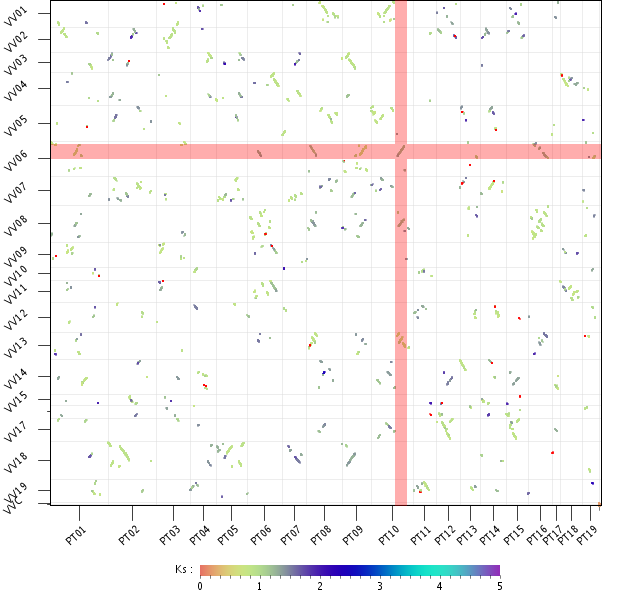
- The WGDotplot here is slightly more difficult to interpret
- For several Vitis segments there are up to 6 poplar segments and for a poplar segment there are up to 3 Vitis segments
- The Ks value shows that for each vitis segment only 2 of the 6 segments have a recent Ks (most recent Ks in the plot is due to the speciation)
- For each poplar region there is only a single Vitis region with the most recent Ks
- This means there was in fact a poplar-specific genome doubling
- Everything else can be explained by the genome triplication that happened before the speciation. Check out the advanced tutorial for more examples like this.
Additional exercises: Uncovering the paleo-hexaploidization
- You should find that the duplication event is shared. Though here it's a triplication (or hexaploidization) instead of a more recent duplication
- With the WGDotplot inspect these combinations
- Papaya-Papaya: Ks is ~1.5
- Vitis-Vitis: Ks is ~ 1
- Papaya-Vitis: Ks < 1
- Conclusion: As the Ks from the speciation is smaller then the duplication's Ks, the speciation is more recent and thus the duplication is shared. Note that these genomes evolve at very different rates, the Ks for the speciation can be higher then the duplication in the slowest evolving genome. Therefore if the Ks of the speciation is lower then one of the duplications it's safe to infer the duplication is shared