Phaeodactylum tricornutum
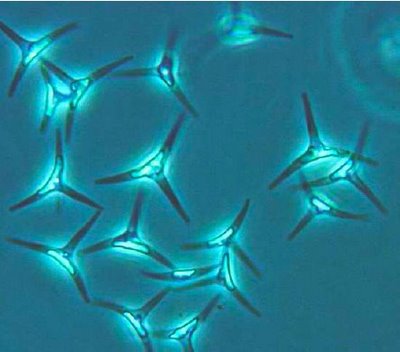
Phaeodactylum tricornutum is a diatom. It is the only species in the genus Phaeodactylum. Unlike other diatoms P. tricornutum can exist in different morphotypes (fusiform, triradiate, and oval), and changes in cell shape can be stimulated by environmental conditions. This feature can be used to explore the molecular basis of cell shape control and morphogenesis. Unlike most diatoms P. tricornutum can grow in the absence of silicon, and the biogenesis of silicified frustules is facultative. This provides opportunities for experimental exploration of silicon-based nanofabrication in diatoms.
Info and Links
| Source | ASM15095v2 |
| PLAZA identifier | ptri |
| NCBI link | Phaeodactylum tricornutum |
| Mitochondrion | HQ840789 |
| Chloroplast | EF067920 |
Toolbox



Annotations
Page 1 of 609, showing 20 records out of 12178 total, starting on record 1, ending on 20
| Gene | Transcript | Start | Stop | Strand | Chr | Type |
|---|---|---|---|---|---|---|
| PTI_00G00010 | PTI_00G00010.1 | 11 | 403 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.4 | coding |
| PTI_00G00020 | PTI_00G00020.1 | 2 | 397 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.5 | coding |
| PTI_00G00030 | PTI_00G00030.1 | 176 | 1984 | - | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.6 | coding |
| PTI_00G00040 | PTI_00G00040.1 | 221 | 481 | - | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.7 | coding |
| PTI_00G00050 | PTI_00G00050.1 | 1217 | 1615 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.7 | coding |
| PTI_00G00060 | PTI_00G00060.1 | 44 | 781 | - | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.8 | coding |
| PTI_00G00070 | PTI_00G00070.1 | 992 | 2059 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.9 | coding |
| PTI_00G00080 | PTI_00G00080.1 | 636 | 839 | - | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.10 | coding |
| PTI_00G00090 | PTI_00G00090.1 | 1438 | 2582 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.11 | coding |
| PTI_00G00100 | PTI_00G00100.1 | 376 | 2382 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.12 | coding |
| PTI_00G00110 | PTI_00G00110.1 | 14 | 666 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.13 | coding |
| PTI_00G00120 | PTI_00G00120.1 | 2 | 664 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.14 | coding |
| PTI_00G00130 | PTI_00G00130.1 | 1543 | 2385 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.14 | coding |
| PTI_00G00140 | PTI_00G00140.1 | 3197 | 4111 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.14 | coding |
| PTI_00G00150 | PTI_00G00150.1 | 20 | 619 | - | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.15 | coding |
| PTI_00G00160 | PTI_00G00160.1 | 942 | 1473 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.15 | coding |
| PTI_00G00170 | PTI_00G00170.1 | 2227 | 2487 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.15 | coding |
| PTI_00G00180 | PTI_00G00180.1 | 3261 | 4130 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.15 | coding |
| PTI_00G00190 | PTI_00G00190.1 | 74 | 3997 | - | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.16 | coding |
| PTI_00G00200 | PTI_00G00200.1 | 25 | 219 | + | contig_tricornutum.ASM15095v2.26.nonchromosomal.dat.17 | coding |
 Expansion_plot
Expansion_plot Gene family finder
Gene family finder Synteny plot
Synteny plot Ks-graphs
Ks-graphs Skyline plot
Skyline plot WGDotplot
WGDotplot WGMapping
WGMapping BLAST
BLAST Workbench
Workbench