| IGR sequence: | igr2102#chr2#x1=10621377#l=6616 |
|---|---|
| Mature miRNA sequence: | CUGACAGAAGAGAGUGAGCACACA |
| encoded miRNA: | MIR63 |
| Precursor location: | 3561 - 3644 (negative strand) |
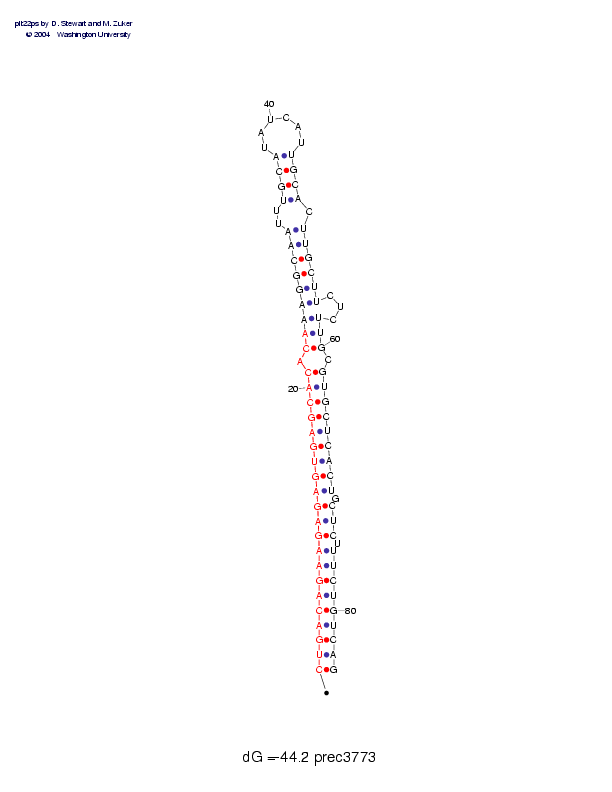
| precursor length: | 84 (34 basepairs) |
| MIR position: | 1 - 24 (3621 - 3644) |
| MIR length: | 24 (23 paired bases) |
| miRNA location TIGR v3: | chr2:10624937<10625020 |
| miRNA location TIGR v5: | chr2:10683550<10683633 |
| Folding energy: | -44.20 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR156a |
| Belongs to miRNAs-targets cluster: | cluster019 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** ********** ****
CUGACAGAAG AGAGUGAGCA CACAAAGGCA AUUUGCAUAU CAUUGCACUU GCUUCUCUUG 60
(((((((((( (((((((((( (-(((((((( (--((((___ ___))))-)) ))))---)))
CGUGCUCACU GCUCUUUCUG UCAG 84
-))))))))) -)))-))))) ))))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At1g53160.1 | 68408.m05523 transcription factor -related | 3 | 8 |
| 2 | At1g69170.1 | 68408.m07265 squamosa-promoter binding protein -related | 4 | 6 |
| 3 | At2g33810.1 | 68409.m03749 squamosa-promoter binding protein -related | 4 | 4 |
| 4 | At2g42200.1 | 68409.m05466 squamosa-promoter binding protein -related | 4 | 8 |
| 5 | At2g42200.2 | 68409.m05467 squamosa-promoter binding protein -related | 4 | 8 |
| 6 | At3g17480.1 | 68410.m02012 F-box protein family | 4 | 1 |
| 7 | At3g57920.1 | 68410.m05967 squamosa promoter-binding protein homolog | 4 | 8 |
| 8 | At5g50570.1 | 68412.m07940 expressed protein | 4 | 8 |
| 9 | At5g50570.2 | 68412.m07941 expressed protein | 4 | 8 |
| 10 | At5g50670.1 | 68412.m05668 expressed protein | 4 | 8 |
Rice homologs
- gnl|BL_ORD_ID|2024_146440+146551_89-112
- gnl|BL_ORD_ID|2024_146440-146551_1-24
- gnl|BL_ORD_ID|2021_81254-81365_1-24
- gnl|BL_ORD_ID|2021_81254+81365_89-112
- gnl|BL_ORD_ID|490_72534-72621_1-24
- gnl|BL_ORD_ID|490_72538-72621_1-24
- gnl|BL_ORD_ID|490_72534+72621_65-88
- gnl|BL_ORD_ID|490_72532+72621_67-90
- gnl|BL_ORD_ID|490_72149-72241_1-24
- gnl|BL_ORD_ID|490_72149+72241_70-93
- gnl|BL_ORD_ID|490_72146+72241_73-96
- gnl|BL_ORD_ID|457_25563-25650_1-24
- gnl|BL_ORD_ID|457_25567-25650_1-24
- gnl|BL_ORD_ID|457_25563+25650_65-88
- gnl|BL_ORD_ID|457_25561+25650_67-90
- gnl|BL_ORD_ID|457_25178-25270_1-24
- gnl|BL_ORD_ID|457_25178+25270_70-93
- gnl|BL_ORD_ID|457_25175+25270_73-96