| IGR sequence: | igr4069#chr3#x1=19080185#l=1539 |
|---|---|
| Mature miRNA sequence: | GAUUGAGCCGCGCCAAUAUCUC |
| encoded miRNA: | MIR30b |
| Precursor location: | 1255 - 1348 (positive strand) |
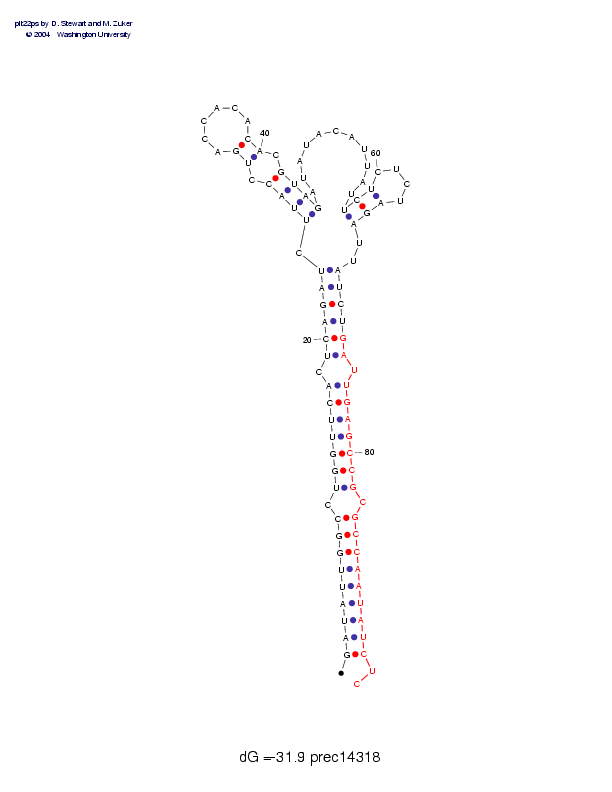
| precursor length: | 94 (31 basepairs) |
| MIR position: | 73 - 94 (1327 - 1348) |
| MIR length: | 22 (18 paired bases) |
| miRNA location TIGR v3: | chr3:19081439>19081532 |
| miRNA location TIGR v5: | chr3:19084429>19084522 |
| Folding energy: | -31.90 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR171 |
| Belongs to miRNAs-targets cluster: | cluster023 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
GAUAUUGGCC UGGUUCACUC AGAUCUUACC UGACCACACA CGUAGAUAUA CAUUAUUCUC 60
(((((((((- (((((((-(( ((((,<<<<- <<______>> ->>>>,,,,, ,,,,,,<<<_
******** ********** ****
UCUAGAUUAU CUGAUUGAGC CGCGCCAAUA UCUC 94
___>>>,,)) ))))-))))) ))-))))))) ))::
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At2g45160.1 | 68409.m05078 scarecrow transcription factor family | 1 | 7 |
| 2 | At3g60630.1 | 68410.m06267 scarecrow transcription factor family | 1 | 7 |
| 3 | At4g00150.1 | 68411.m00015 scarecrow-like transcription factor 6 (SCL6) | 1 | 7 |
Rice homologs
- gnl|BL_ORD_ID|263_84399-84502_1-22
- gnl|BL_ORD_ID|263_84399+84502_83-104