| IGR sequence: | igr2356#chr3#x1=9872012#l=10062 |
|---|---|
| Mature miRNA sequence: | GGUAGCCAAGGAUGACUUGCCU |
| encoded miRNA: | MIR43 |
| Precursor location: | 4804 - 4933 (negative strand) |
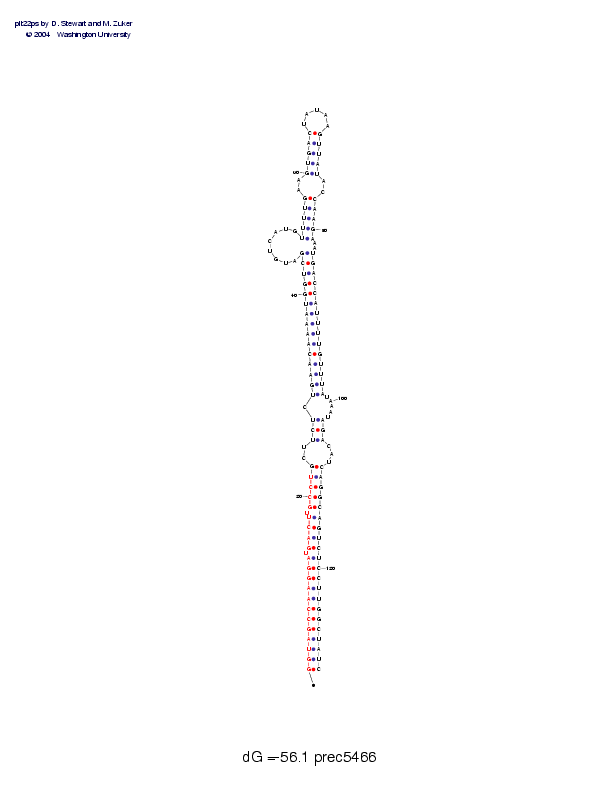
| precursor length: | 130 (49 basepairs) |
| MIR position: | 1 - 22 (4912 - 4933) |
| MIR length: | 22 (20 paired bases) |
| miRNA location TIGR v3: | chr3:9876815<9876944 |
| miRNA location TIGR v5: | chr3:9876805<9876934 |
| Folding energy: | -56.10 |
| BLAST hit against RFAM Atha miRNAs: | none |
| Belongs to miRNAs-targets cluster: | cluster006 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** ********** **
GGUAGCCAAG GAUGACUUGC CUGCUUCUCU GAACAAAAUG GUCGAUGUCA UGUUUUGAAG 60
(((((((((( ((-((((-(( (((--(((-( (((((((((( ((((------ --(((((--(
UGACUAUAAG UUAUACCAAG AAAUGACCAU UUUGUUUAUA AAUAGACAUC AGGCAGUCUC 120
((((_____) ))))--)))) )--))))))) ))))))))-- ---)))---) ))))))))))
CUUGGCUAUC 130
))))))))))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At1g17590.1 | 68408.m01959 transcription factor -related | 3 | 10 |
| 2 | At1g54160.1 | 68408.m05664 CCAAT-binding factor B subunit -related | 3 | 8 |
| 3 | At1g72830.1 | 68408.m07728 CCAAT-binding factor B subunit homolog -related | 3 | 8 |
Rice homologs
- gnl|BL_ORD_ID|2154_158028-158181_1-22
- gnl|BL_ORD_ID|2154_158030+158181_131-152
- gnl|BL_ORD_ID|323_84895+85046_131-152
- gnl|BL_ORD_ID|323_84893-85046_1-22