| IGR sequence: | igr1679#chr4#x1=8848676#l=7609 |
|---|---|
| Mature miRNA sequence: | UGCCUGGCUCCCUGUAUGCCAC |
| encoded miRNA: | MIR75 |
| Precursor location: | 5318 - 5398 (positive strand) |
| precursor length: | 81 (29 basepairs) |
| MIR position: | 1 - 22 (5318 - 5339) |
| MIR length: | 22 (19 paired bases) |
| miRNA location TIGR v3: | chr4:8853993>8854073 |
| miRNA location TIGR v5: | chr4:9888999>9889079 |
| Folding energy: | -34.50 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR160b |
| Belongs to miRNAs-targets cluster: | cluster017 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** ********** **
UGCCUGGCUC CCUGUAUGCC ACAAGAAAAC AUCGAUUUAG UUUCAAAAUC GAUCACUAGU 60
:((-(((((- (((((((((( ((-((----- ((((((((__ _____))))) )))--))-))
GGCGUACAGA GUAGUCAAGC A 81
)))))))))- )-)))))-)) :
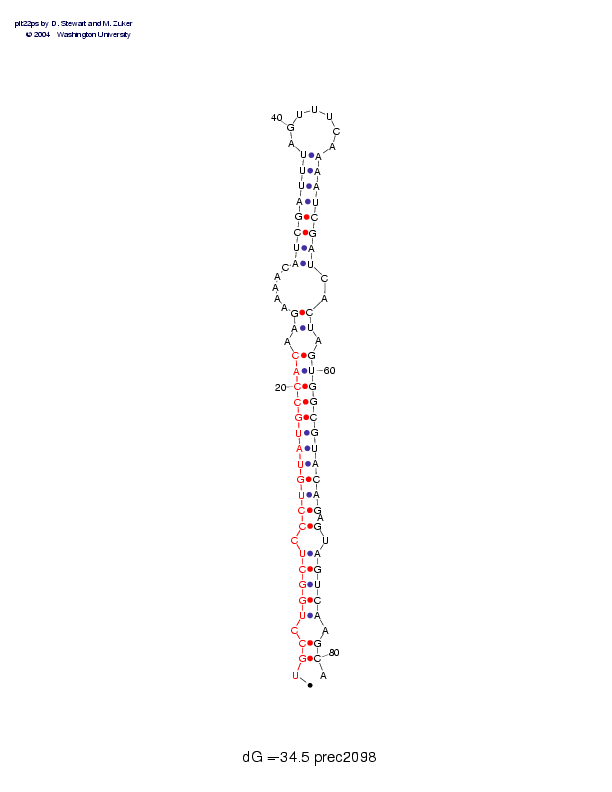
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At1g77850.1 | 68408.m08325 transcriptional factor B3 family | 2 | 7 |
| 2 | At2g28350.1 | 68409.m03117 auxin response transcription factor | 3 | 4 |
Rice homologs
- gnl|BL_ORD_ID|46_2746+2875_1-22
- gnl|BL_ORD_ID|16_152197+152326_1-22