| IGR sequence: | igr4444#chr3#x1=20692045#l=9635 |
|---|---|
| Mature miRNA sequence: | AUCCAAAGGGAUCGCAUUGAUCCU |
| encoded miRNA: | MIR10 |
| Precursor location: | 7607 - 7748 (positive strand) |
| precursor length: | 142 (47 basepairs) |
| MIR position: | 1 - 24 (7607 - 7630) |
| MIR length: | 24 (22 paired bases) |
| miRNA location TIGR v3: | chr3:20699651>20699792 |
| miRNA location TIGR v5: | chr3:20702635>20702776 |
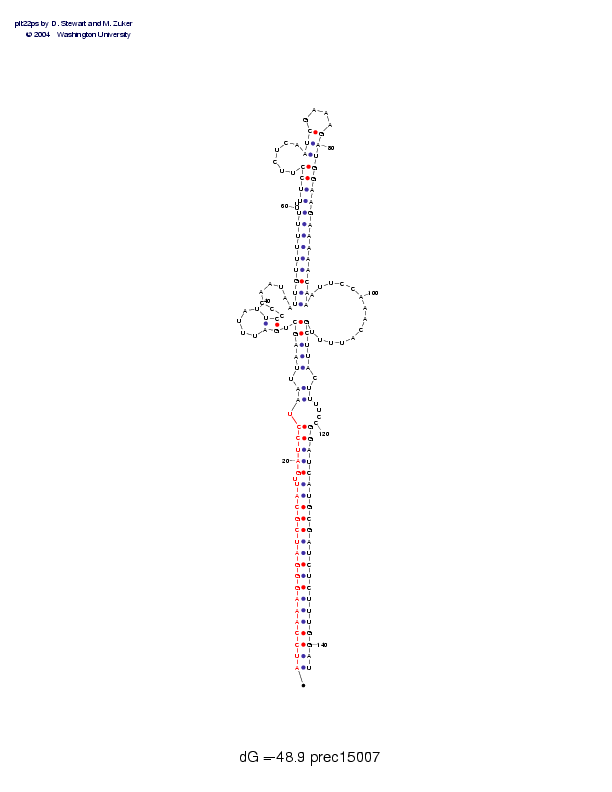
| Folding energy: | -48.90 |
| BLAST hit against RFAM Atha miRNAs: | none |
| Belongs to miRNAs-targets cluster: | cluster009 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** ********** ****
AUCCAAAGGG AUCGCAUUGA UCCUAAUUAA GCUGAUUUAU UCCCCAAUAA UUGUUUUUUU 60
(((((((((( (((((((-(( (((-((-((( ((,<<_____ >>,,,,,,,, <<<<<<<<<-
UUUCCUUCUC AAUCGAAAGA UGGAAGAAAA ACAAAUUCCA AACAUUUUGC UUACUUUUCC 120
-<<<<----- -<<<____>> >>>>>>>>>> >>>>,,,,,, ,,,,,,,,)) )))-))----
GGAUCAUGCG AUCUCUUUGG AU 142
)))))))))) )))))))))) ))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At1g12820.1 | 68408.m01345 transport inhibitor response 1 (TIR1), putative | 3 | 2 |
| 2 | At3g26810.1 | 68410.m03071 transport inhibitor response 1 (TIR1), putative | 3 | 2 |
| 3 | At3g62980.1 | 68410.m06538 transport inhibitor response 1 (TIR1), AtFBL1 | 4 | 1 |
| 4 | At4g03190.1 | 68411.m00397 F-box protein GRR1-like protein 1, AtFBL18 | 4 | 1 |
Rice homologs
- gnl|BL_ORD_ID|517_37281-37376_73-96
- gnl|BL_ORD_ID|517_37281+37376_1-24
- gnl|BL_ORD_ID|464_91815-91910_73-96
- gnl|BL_ORD_ID|464_91815+91910_1-24