| IGR sequence: | igr4079#chr2#x1=18941708#l=2762 |
|---|---|
| Mature miRNA sequence: | UUGGAUUGAAGGGAGCUCCU |
| encoded miRNA: | MIR88 |
| Precursor location: | 1395 - 1599 (positive strand) |
| precursor length: | 205 (73 basepairs) |
| MIR position: | 186 - 205 (1580 - 1599) |
| MIR length: | 20 (18 paired bases) |
| miRNA location TIGR v3: | chr2:18943102>18943306 |
| miRNA location TIGR v5: | chr2:19001715>19001919 |
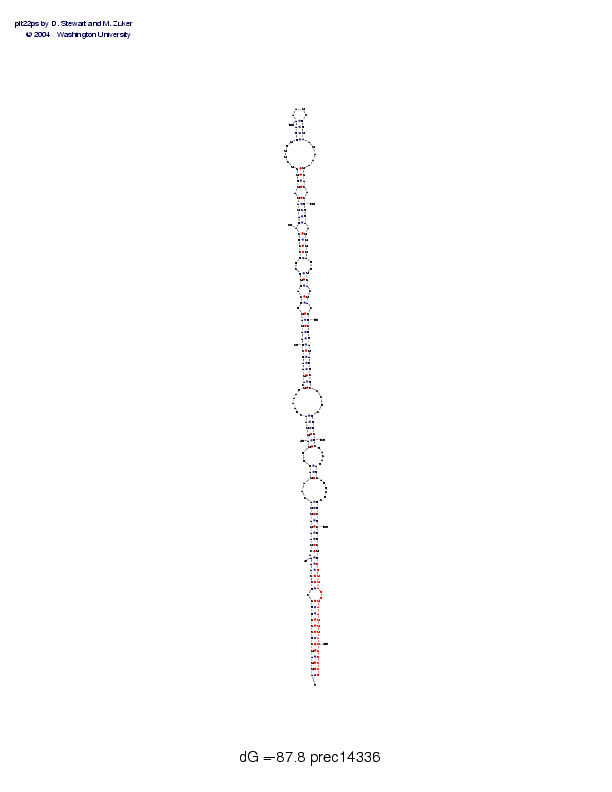
| Folding energy: | -87.80 |
| BLAST hit against RFAM Atha miRNAs: | none |
| Belongs to miRNAs-targets cluster: | cluster034 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
AGGAGCUCCC UUCCUCCAAA ACGAAGAGGA CAAGAUUUGA GGAACUAAAA UGCAGAAUCU 60
(((((((((( (((-(((((( -((((((((( ---(((--(( ((((------ -(-(((((((
AAGAGUUCAU GUCUUCCUCA UAGAGAGUGC GCGGUGUUAA AAGCUUGAAG AAAGCACACU 120
(((((-((-( ((--(((((- (((((-(((( ------(((( ____))))-- ---))))-))
UUAAGGGGAU UGCACGACCU CUUAGAUUCU CCCUCUUUCU CUACAUAUCA UUCUCUUCUC 180
)))-)))))- -)))-))-)) )))))))))) )----))))) )-----)))- -----)))))
***** ********** *****
UUCGUUUGGA UUGAAGGGAG CUCCU 205
)))))))))) --)))))))) )))))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At2g26950.1 | 68409.m02933 myb family transcription factor | 2 | 1 |
| 2 | At3g11440.1 | 68410.m01260 myb family transcription factor | 2 | 1 |
| 3 | At3g33076.1 | 68410.m03915 retrotransposon gag protein family | 2 | 1 |
| 4 | At5g06100.1 | 68412.m00630 myb family transcription factor | 2 | 1 |
| 5 | At5g06100.2 | 68412.m00631 myb family transcription factor | 2 | 1 |
| 6 | At5g55020.1 | 68412.m06200 expressed protein | 2 | 1 |
Rice homologs
- gnl|BL_ORD_ID|479_23793-23963_1-20
- gnl|BL_ORD_ID|479_23793+23963_152-171