| IGR sequence: | igr3768#chr5#x1=18615936#l=2646 |
|---|---|
| Mature miRNA sequence: | UGCCUGGCUCCCUGUAUGCCAC |
| encoded miRNA: | MIR75 |
| Precursor location: | 1332 - 1412 (negative strand) |
| precursor length: | 81 (30 basepairs) |
| MIR position: | 1 - 22 (1391 - 1412) |
| MIR length: | 22 (19 paired bases) |
| miRNA location TIGR v3: | chr5:18617267<18617347 |
| miRNA location TIGR v5: | chr5:19026325<19026405 |
| Folding energy: | -43.80 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR160c |
| Belongs to miRNAs-targets cluster: | cluster017 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** ********** **
UGCCUGGCUC CCUGUAUGCC ACGAGUGGAU ACCGAUUUUG GUUUUAAAAU CGGCUGCCGG 60
:((-(((((( (-(((((((( ((--(--(-- -((((((((( ____)))))) ))))--)--)
UGGCGUACAA GGAGUCAAGC A 81
)))))))))- )))))))-)) :
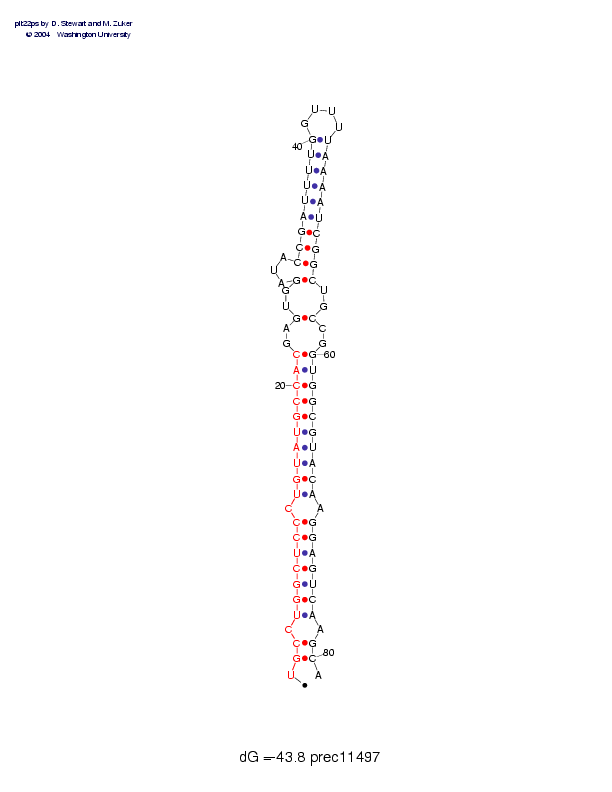
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At1g77850.1 | 68408.m08325 transcriptional factor B3 family | 2 | 7 |
| 2 | At2g28350.1 | 68409.m03117 auxin response transcription factor | 3 | 4 |
Rice homologs
- gnl|BL_ORD_ID|1495_131987+132068_61-82
- gnl|BL_ORD_ID|1495_131987-132068_1-22
- gnl|BL_ORD_ID|1461_56381+56462_61-82
- gnl|BL_ORD_ID|1461_56381-56462_1-22
- gnl|BL_ORD_ID|1294_49693-49779_1-22
- gnl|BL_ORD_ID|727_84855-84941_1-22