| IGR sequence: | igr280#chr5#x1=1185020#l=7141 |
|---|---|
| Mature miRNA sequence: | GAAUCUUGAUGAUGCUGCAUC |
| encoded miRNA: | MIR18 |
| Precursor location: | 3192 - 3282 (negative strand) |
| precursor length: | 91 (33 basepairs) |
| MIR position: | 71 - 91 (3192 - 3212) |
| MIR length: | 21 (17 paired bases) |
| miRNA location TIGR v3: | chr5:1188211<1188301 |
| miRNA location TIGR v5: | chr5:1188211<1188301 |
| Folding energy: | -35.90 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR172a2 |
| Belongs to miRNAs-targets cluster: | cluster011 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
GGCGCAGCAC CAUUAAGAUU CACAUGGAAA UUGAUAAAUA CCCUAAAUUA GGGUUUUGAU 60
:::((((((- (((((((((( (-((((---( (---((((-( (((((___)) ))))))))--
********** ********** *
AUGUAUAUGA GAAUCUUGAU GAUGCUGCAU C 91
))---))))- )))))))))) )-)))))):: :
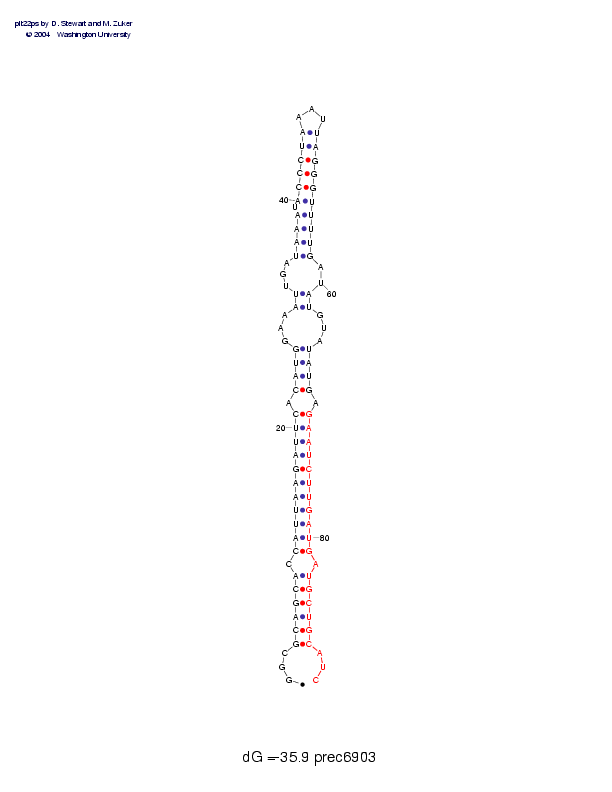
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At4g36920.1 | 68411.m04757 floral homeotic protein APETALA2 | 2 | 3 |
| 2 | At5g60120.1 | 68412.m06823 AP2 domain transcription factor, putative | 2 | 3 |
Rice homologs
- gnl|BL_ORD_ID|1572_16982+17201_1-21
- gnl|BL_ORD_ID|1572_16982-17201_200-220
- gnl|BL_ORD_ID|526_109837+110056_1-21
- gnl|BL_ORD_ID|526_109837-110056_200-220