| IGR sequence: | igr5124#chr5#x1=24569732#l=3135 |
|---|---|
| Mature miRNA sequence: | UGCCAAAGGAGAGUUGCCCU |
| encoded miRNA: | MIR16 |
| Precursor location: | 933 - 1024 (positive strand) |
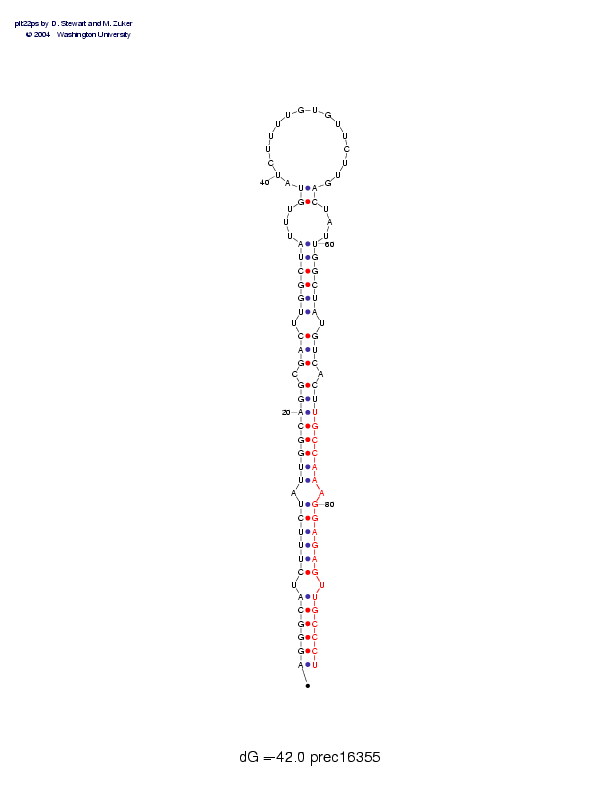
| precursor length: | 92 (31 basepairs) |
| MIR position: | 73 - 92 (1005 - 1024) |
| MIR length: | 20 (18 paired bases) |
| miRNA location TIGR v3: | chr5:24570664>24570755 |
| miRNA location TIGR v5: | chr5:24979722>24979813 |
| Folding energy: | -41.99 |
| BLAST hit against RFAM Atha miRNAs: | none |
| Belongs to miRNAs-targets cluster: | cluster020 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
AGGGCAUCUU UCUAUUGGCA GGCGACUUGG CUAUUUGUAU CUUUUGUGUU CUUGACUAUU 60
((((((-((( (((-(((((( ((-(((-((( (((---((__ __________ ____))---)
******** ********** **
GGCUAUGUCA CUUGCCAAAG GAGAGUUGCC CU 92
)))))-)))- ))))))))-) )))))-)))) ))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At2g33770.1 | 68409.m03744 ubiquitin-conjugating enzyme family | 2 | 5 |
| 2 | At2g33770.1 | 68409.m03744 ubiquitin-conjugating enzyme family | 2 | 5 |
| 3 | At2g33770.1 | 68409.m03744 ubiquitin-conjugating enzyme family | 2 | 5 |
| 4 | At2g33770.1 | 68409.m03744 ubiquitin-conjugating enzyme family | 2 | 5 |
Rice homologs
- gnl|BL_ORD_ID|2430_58466+58549_65-84
- gnl|BL_ORD_ID|2430_58466-58549_1-20