| IGR sequence: | igr4203#chr2#x1=19467319#l=3008 |
|---|---|
| Mature miRNA sequence: | GUUGGAGAAGCAGGGCACGUGCA |
| encoded miRNA: | MIR29 |
| Precursor location: | 1907 - 1988 (positive strand) |
| precursor length: | 82 (29 basepairs) |
| MIR position: | 1 - 23 (1907 - 1929) |
| MIR length: | 23 (20 paired bases) |
| miRNA location TIGR v3: | chr2:19469225>19469306 |
| miRNA location TIGR v5: | chr2:19527838>19527919 |
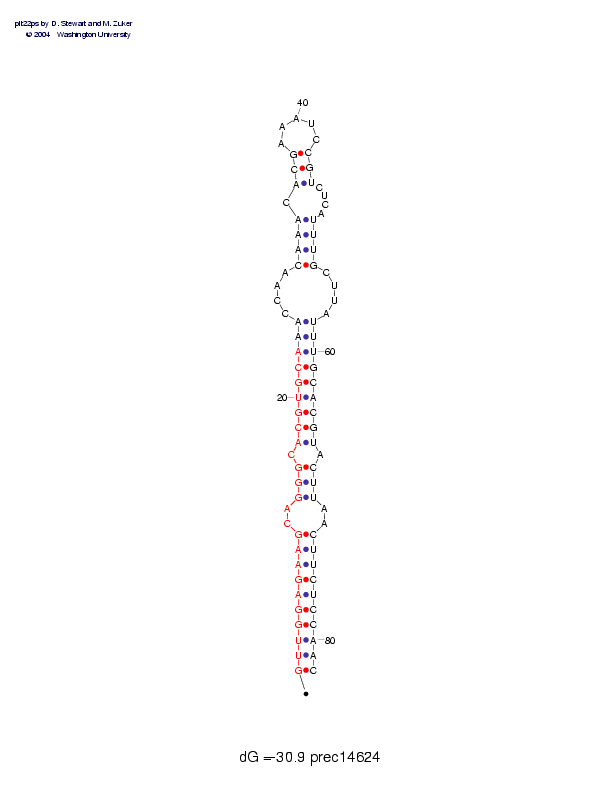
| Folding energy: | -30.90 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR164a |
| Belongs to miRNAs-targets cluster: | cluster026 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** ********** ***
GUUGGAGAAG CAGGGCACGU GCAAACCAAC AAACACGAAA UCCGUCUCAU UUGCUUAUUU 60
(((((((((( --(((-(((( (((((----( (((-(((___ __)))----) )))----)))
GCACGUACUU AACUUCUCCA AC 82
))))))-))) --)))))))) ))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At1g56010.1 | 68408.m05888 NAC1 / No apical meristem (NAM) protein family | 3 | 2 |
| 2 | At1g56010.2 | 68408.m05889 NAC1 / No apical meristem (NAM) protein family | 3 | 2 |
| 3 | At5g07680.1 | 68412.m00813 NAM (no apical meristem)-related protein | 2 | 2 |
| 4 | At5g07680.2 | 68412.m00814 NAM (no apical meristem)-related protein | 2 | 2 |
| 5 | At5g61430.1 | 68412.m06973 NAM, no apical meristem, - like protein | 2 | 2 |
Rice homologs
- gnl|BL_ORD_ID|968_113708+113833_1-23
- gnl|BL_ORD_ID|968_113708-113833_104-126