| IGR sequence: | igr2928#chr4#x1=14377290#l=6844 |
|---|---|
| Mature miRNA sequence: | UGUGUGCUCACUCUCUUCUGUCAG |
| encoded miRNA: | MIR63a |
| Precursor location: | 3138 - 3222 (positive strand) |
| precursor length: | 85 (34 basepairs) |
| MIR position: | 62 - 85 (3199 - 3222) |
| MIR length: | 24 (24 paired bases) |
| miRNA location TIGR v3: | chr4:14380427>14380511 |
| miRNA location TIGR v5: | chr4:15415433>15415517 |
| Folding energy: | -45.90 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR156c |
| Belongs to miRNAs-targets cluster: | none |
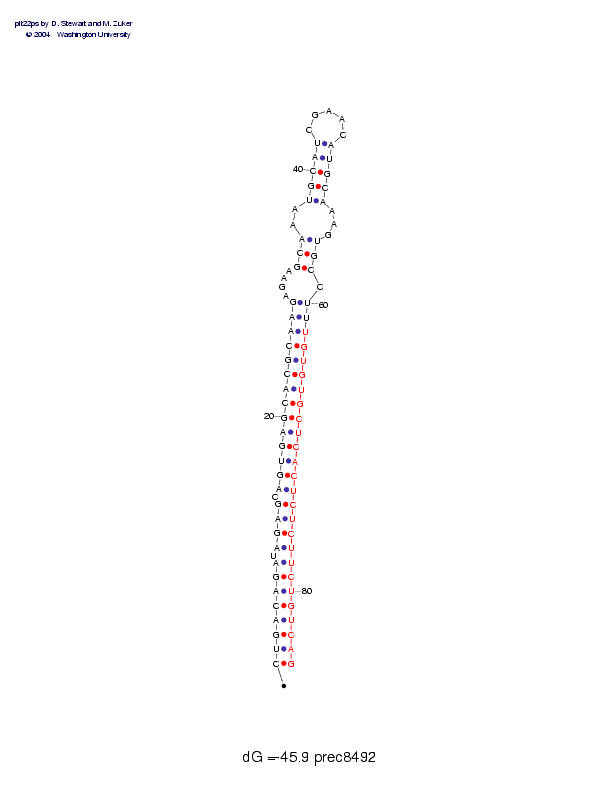
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
CUGACAGAUA GAGCAGUGAG CACGCAAGAG AAGCAAAUGC AUCGAACAUG CAAAGUGCCU 60
((((((((-( (((-(((((( ((((((((-- --(((--((( ((_____))) ))---)))-)
********* ********** *****
UUGUGUGCUC ACUCUCUUCU GUCAG 85
)))))))))) )))))))))) )))))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|2024_146440+146551_89-112
- gnl|BL_ORD_ID|2024_146440-146551_1-24
- gnl|BL_ORD_ID|2021_81254-81365_1-24
- gnl|BL_ORD_ID|2021_81254+81365_89-112
- gnl|BL_ORD_ID|490_72534-72621_1-24
- gnl|BL_ORD_ID|490_72538-72621_1-24
- gnl|BL_ORD_ID|490_72534+72621_65-88
- gnl|BL_ORD_ID|490_72532+72621_67-90
- gnl|BL_ORD_ID|490_72149-72241_1-24
- gnl|BL_ORD_ID|490_72149+72241_70-93
- gnl|BL_ORD_ID|490_72146+72241_73-96
- gnl|BL_ORD_ID|457_25563-25650_1-24
- gnl|BL_ORD_ID|457_25567-25650_1-24
- gnl|BL_ORD_ID|457_25563+25650_65-88
- gnl|BL_ORD_ID|457_25561+25650_67-90
- gnl|BL_ORD_ID|457_25178-25270_1-24
- gnl|BL_ORD_ID|457_25178+25270_70-93
- gnl|BL_ORD_ID|457_25175+25270_73-96