| IGR sequence: | igr2848#chr4#x1=14039889#l=4299 |
|---|---|
| Mature miRNA sequence: | GUGCUCACCUCUCUUUCUGUCAG |
| encoded miRNA: | MIR61 |
| Precursor location: | 56 - 137 (positive strand) |
| precursor length: | 82 (33 basepairs) |
| MIR position: | 60 - 82 (115 - 137) |
| MIR length: | 23 (21 paired bases) |
| miRNA location TIGR v3: | chr4:14039944>14040025 |
| miRNA location TIGR v5: | chr4:15074950>15075031 |
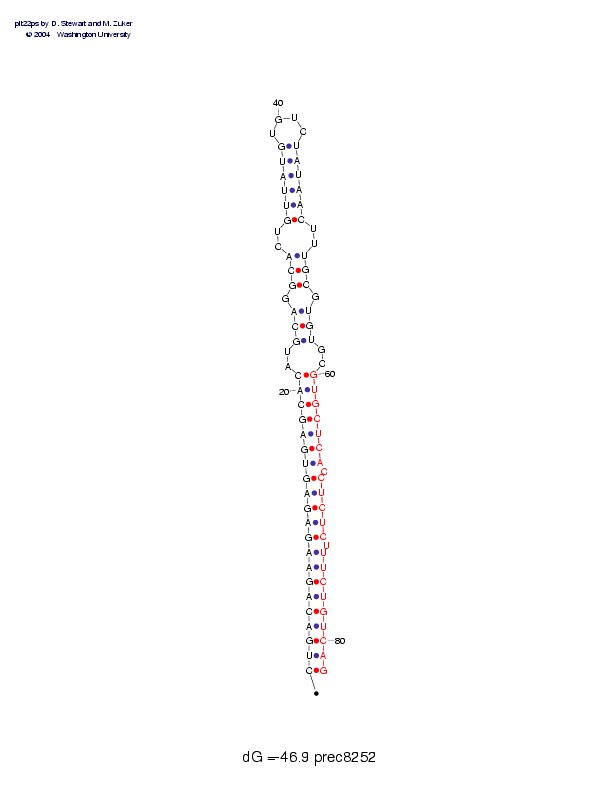
| Folding energy: | -46.90 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR156b |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
*
CUGACAGAAG AGAGUGAGCA CAUGCAGGCA CUGUUAUGUG UCUAUAACUU UGCGUGUGCG 60
(((((((((( (((((((((( (--(((-((( --((((((__ __))))))-- )))-)))--)
********** ********** **
UGCUCACCUC UCUUUCUGUC AG 82
))))))-))) ))-))))))) ))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|668_88196+88333_116-138
- gnl|BL_ORD_ID|668_88196-88333_1-23