| IGR sequence: | igr2385#chr2#x1=11888418#l=10031 |
|---|---|
| Mature miRNA sequence: | GUGGCAUCAUCAAGAUUCACA |
| encoded miRNA: | MIR65 |
| Precursor location: | 2967 - 3058 (negative strand) |
| precursor length: | 92 (34 basepairs) |
| MIR position: | 1 - 21 (3038 - 3058) |
| MIR length: | 21 (18 paired bases) |
| miRNA location TIGR v3: | chr2:11891384<11891475 |
| miRNA location TIGR v5: | chr2:11949997<11950088 |
| Folding energy: | -40.30 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR172a1 |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** ********** *
GUGGCAUCAU CAAGAUUCAC AUCUGUUGAU GGACGGUGGU GAUUCACUCU CCACAAAGUU 60
(--((((((( ((((((((-( ((,,,<<<-< <<<-<<<<<_ ___>>>>>-> >>>>>>,,<<
CUCUAUGAAA AUGAGAAUCU UGAUGAUGCU GC 92
<_____>>>, )))-)))))) )))))))))- -)
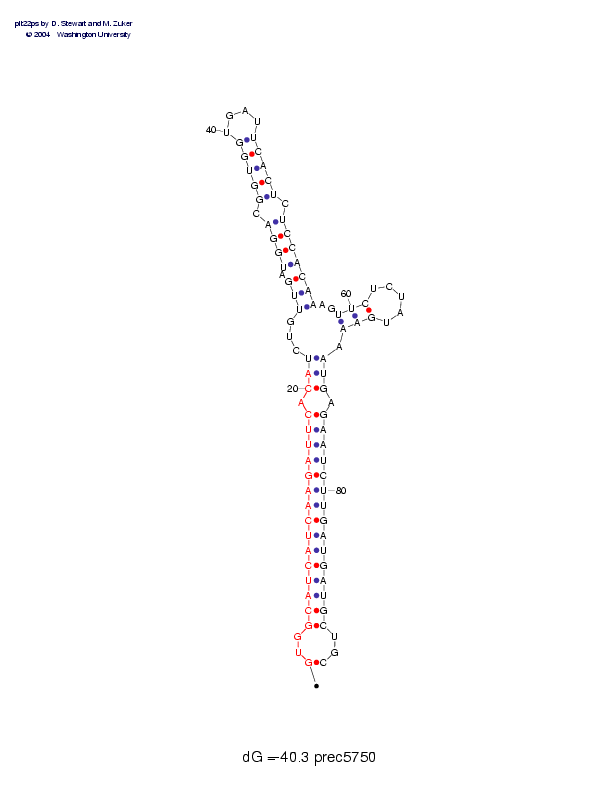
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1572_16985-17198_1-21
- gnl|BL_ORD_ID|526_109840-110053_1-21