| IGR sequence: | igr216#chr2#x1=1039061#l=1163 |
|---|---|
| Mature miRNA sequence: | UGUGUUCUCAGGUCACCCCUUUG |
| encoded miRNA: | MIR1 |
| Precursor location: | 883 - 971 (positive strand) |
| precursor length: | 89 (30 basepairs) |
| MIR position: | 67 - 89 (949 - 971) |
| MIR length: | 23 (20 paired bases) |
| miRNA location TIGR v3: | chr2:1039943>1040031 |
| miRNA location TIGR v5: | chr2:1040943>1041031 |
| Folding energy: | -36.24 |
| BLAST hit against RFAM Atha miRNAs: | none |
| Belongs to miRNAs-targets cluster: | cluster031 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
CAAAGGAGUG GCAUGUGAAC ACAUAUCCUA UGGUUUCUUC AAAUUUCCAU UGAAACCAUU 60
((((((-((( ((-((-(((( (((--((--( (((((((___ __________ _))))))))-
**** ********** *********
GAGUUUUGUG UUCUCAGGUC ACCCCUUUG 89
))----)))) )))-))-))) ))-))))))
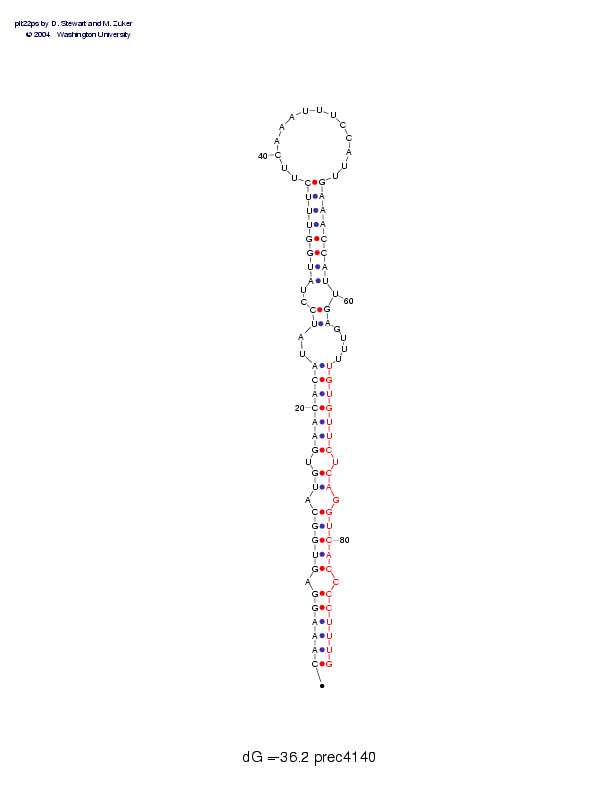
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At1g08830.1 | 68408.m00884 copper/zinc superoxidase dismutase (CSD1) | 3 | 1 |
Rice homologs
- gnl|BL_ORD_ID|1328_130435-130533_1-23
- gnl|BL_ORD_ID|1328_130435+130533_77-99