| IGR sequence: | igr2102#chr2#x1=10621377#l=6616 |
|---|---|
| Mature miRNA sequence: | UGACAGAAAGAGCAGUGAGCACGC |
| encoded miRNA: | MIR68 |
| Precursor location: | 3562 - 3643 (positive strand) |
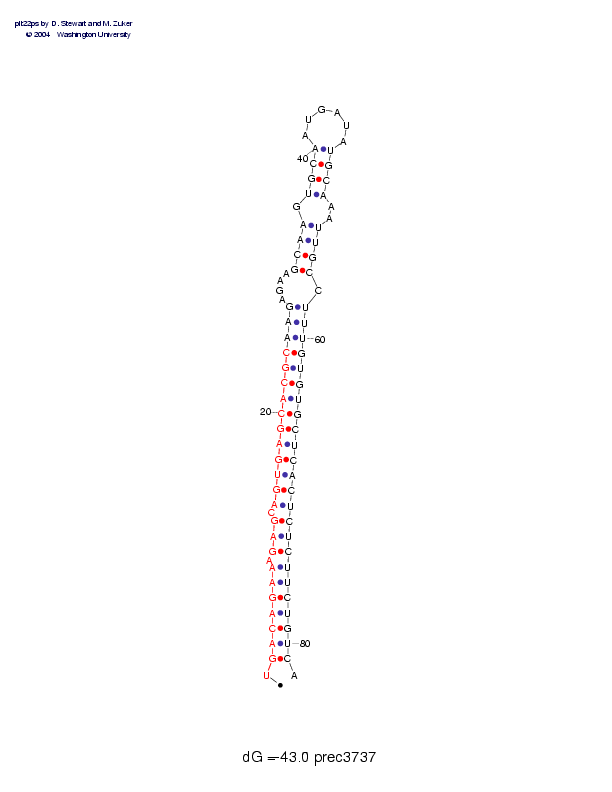
| precursor length: | 82 (32 basepairs) |
| MIR position: | 1 - 24 (3562 - 3585) |
| MIR length: | 24 (21 paired bases) |
| miRNA location TIGR v3: | chr2:10624938>10625019 |
| miRNA location TIGR v5: | chr2:10683551>10683632 |
| Folding energy: | -43.00 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR156a |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** ********** ****
UGACAGAAAG AGCAGUGAGC ACGCAAGAGA AGCAAGUGCA AUGAUAUGCA AAUUGCCUUU 60
:(((((((-( ((-((((((( (((((((--- -((((-(((( ______)))) --))))-)))
GUGUGCUCAC UCUCUUCUGU CA 82
)))))))))) )))))))))) ):
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|2522_50991-51076_63-86
- gnl|BL_ORD_ID|2522_50991+51076_1-24
- gnl|BL_ORD_ID|640_18845-18930_63-86
- gnl|BL_ORD_ID|640_18845+18930_1-24
- gnl|BL_ORD_ID|342_115960-116043_61-84
- gnl|BL_ORD_ID|342_115960+116043_1-24