| IGR sequence: | igr2044#chr5#x1=9109684#l=5223 |
|---|---|
| Mature miRNA sequence: | GUGUGCUCACUCUCUUCUGUCACC |
| encoded miRNA: | MIR17b |
| Precursor location: | 4188 - 4281 (negative strand) |
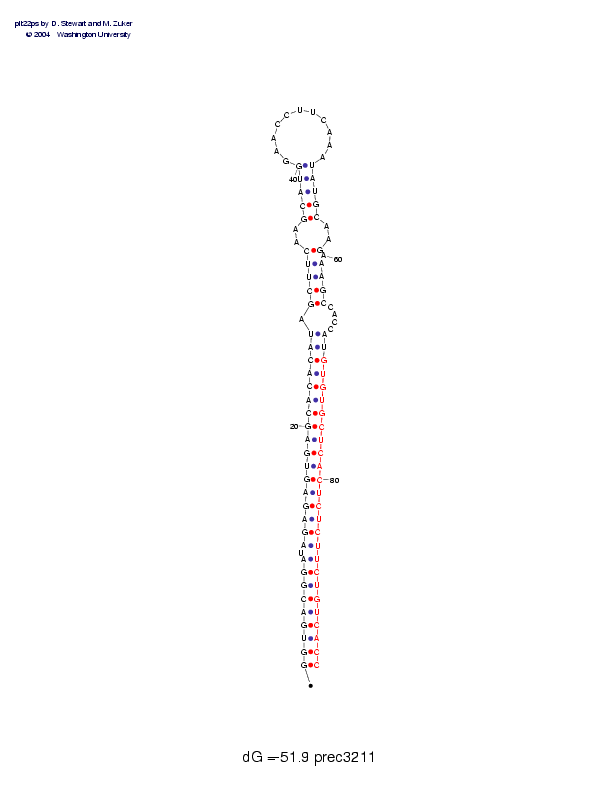
| precursor length: | 94 (36 basepairs) |
| MIR position: | 71 - 94 (4188 - 4211) |
| MIR length: | 24 (24 paired bases) |
| miRNA location TIGR v3: | chr5:9113871<9113964 |
| miRNA location TIGR v5: | chr5:9136127<9136220 |
| Folding energy: | -51.90 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR156f |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
GGUGACGGAU AGAGAGUGAG CACACAUAGC UUCAAGCAUG GAACCUUCAA AUAUGCAAGA 60
(((((((((- (((((((((( (((((((-(( (((--((((( __________ _)))))--)-
********** ********** ****
AAGCCACCAU GUGUGCUCAC UCUCUUCUGU CACC 94
))))----)) )))))))))) )))))))))) ))))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|2522_50927+51016_1-24
- gnl|BL_ORD_ID|2522_50927-51016_67-90
- gnl|BL_ORD_ID|640_18781+18870_1-24
- gnl|BL_ORD_ID|640_18781-18870_67-90