| IGR sequence: | igr1987#chr3#x1=8101048#l=11724 |
|---|---|
| Mature miRNA sequence: | AGAUCAUGCUGGCAGCUUCAUC |
| encoded miRNA: | MIR53 |
| Precursor location: | 7048 - 7152 (negative strand) |
| precursor length: | 105 (38 basepairs) |
| MIR position: | 84 - 105 (7048 - 7069) |
| MIR length: | 22 (19 paired bases) |
| miRNA location TIGR v3: | chr3:8108095<8108199 |
| miRNA location TIGR v5: | chr3:8108095<8108199 |
| Folding energy: | -30.00 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR167a |
| Belongs to miRNAs-targets cluster: | cluster005 |
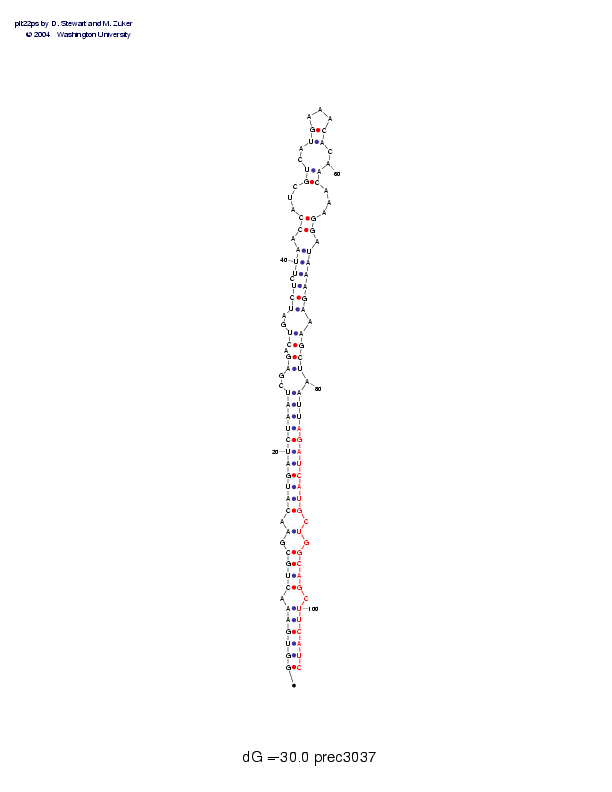
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
GGUGAAACUG CGAACAUGAU CUAAUCGAGA CUGAUCUCUU AACCAUCGUC AUGAAACACA 60
((((((-((( (-(-(((((( (((((--((- ((--(((-(( (-((---((- -((___))--
******* ********** *****
ACAAAGGAUA AAGAAAGCUA AUUAGAUCAU GCUGGCAGCU UCAUC 105
))---))-)) ))))-))))- )))))))))) )-)-))))-) )))))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At3g06870.1 | 68410.m00742 proline-rich protein family | 3 | 1 |
Rice homologs
- gnl|BL_ORD_ID|822_81028-81124_76-97
- gnl|BL_ORD_ID|822_81028+81124_1-22