| IGR sequence: | igr17#chr1#x1=77747#l=4997 |
|---|---|
| Mature miRNA sequence: | CCAGACAACAUUCCCCUCAACUG |
| encoded miRNA: | MIR50 |
| Precursor location: | 1181 - 1293 (positive strand) |
| precursor length: | 113 (36 basepairs) |
| MIR position: | 91 - 113 (1271 - 1293) |
| MIR length: | 23 (18 paired bases) |
| miRNA location TIGR v3: | chr1:78927>79039 |
| miRNA location TIGR v5: | chr1:78927>79039 |
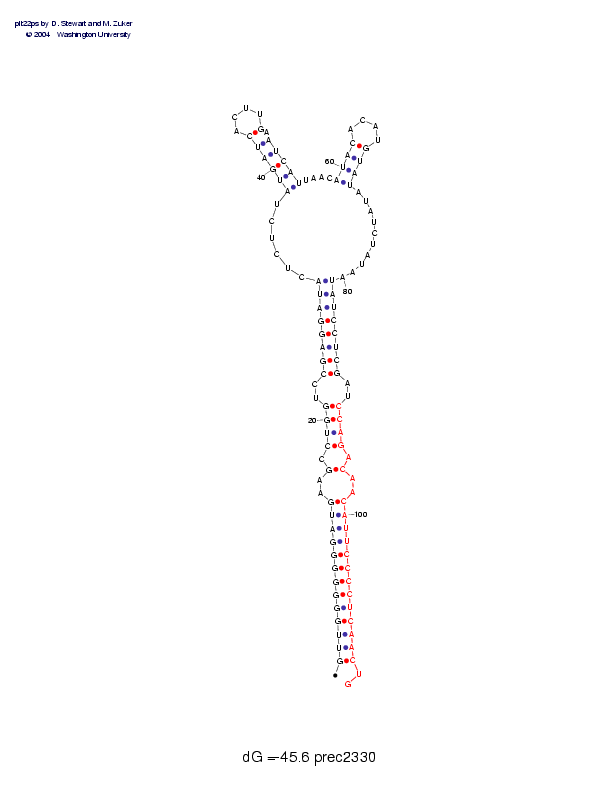
| Folding energy: | -45.60 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR165a |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
GUUGGGGGGG AUGAAGCCUG GUCCGAGGAU ACUCUCUAUG AUCACUUGAA UCAUUAACAU 60
(((((((((( (((--(-((( (--((((((( (,,,,,,<<< <<<____>-> >>>>,,,,<<
********** ********** ***
ACACAUGUAU AUAUCUAUAA UAUCCUCGAU CCAGACAACA UUCCCCUCAA CUG 113
<<____>>>> ,,,,,,,,,, ))))))))-- ))))-)--)) )))))))))) )::
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1553_40795+40909_93-115
- gnl|BL_ORD_ID|1553_40795-40909_1-23