| IGR sequence: | igr1679#chr4#x1=8848676#l=7609 |
|---|---|
| Mature miRNA sequence: | GUGGCAUACAGGGAGCCAGGCAC |
| encoded miRNA: | MIR75d |
| Precursor location: | 5317 - 5398 (negative strand) |
| precursor length: | 82 (30 basepairs) |
| MIR position: | 60 - 82 (5317 - 5339) |
| MIR length: | 23 (20 paired bases) |
| miRNA location TIGR v3: | chr4:8853992<8854073 |
| miRNA location TIGR v5: | chr4:9888998<9889079 |
| Folding energy: | -35.20 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR160b |
| Belongs to miRNAs-targets cluster: | none |
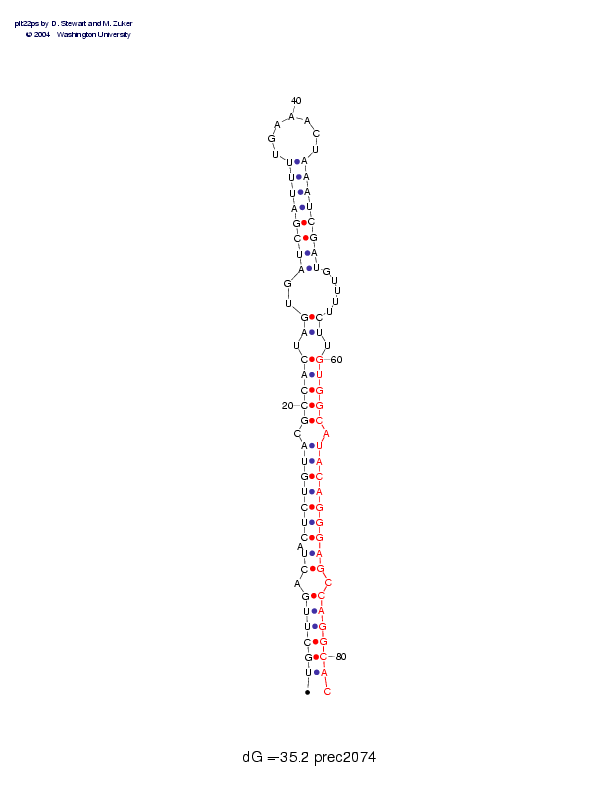
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
*
UGCUUGACUA CUCUGUACGC CACUAGUGAU CGAUUUUGAA ACUAAAUCGA UGUUUUCUUG 60
((((((-((- (((((((-(( (((-((--(( ((((((____ ___))))))) )-----))-)
********** ********** **
UGGCAUACAG GGAGCCAGGC AC 82
))))-))))) ))))-))))) ):
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1294_49758+49846_1-23
- gnl|BL_ORD_ID|727_84919+85007_1-23