| IGR sequence: | igr1679#chr4#x1=8848676#l=7609 |
|---|---|
| Mature miRNA sequence: | GUGGCAUACAGGGAGCCAGGCACG |
| encoded miRNA: | MIR75b |
| Precursor location: | 5316 - 5400 (negative strand) |
| precursor length: | 85 (30 basepairs) |
| MIR position: | 62 - 85 (5316 - 5339) |
| MIR length: | 24 (20 paired bases) |
| miRNA location TIGR v3: | chr4:8853991<8854075 |
| miRNA location TIGR v5: | chr4:9888997<9889081 |
| Folding energy: | -35.50 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR160b |
| Belongs to miRNAs-targets cluster: | none |
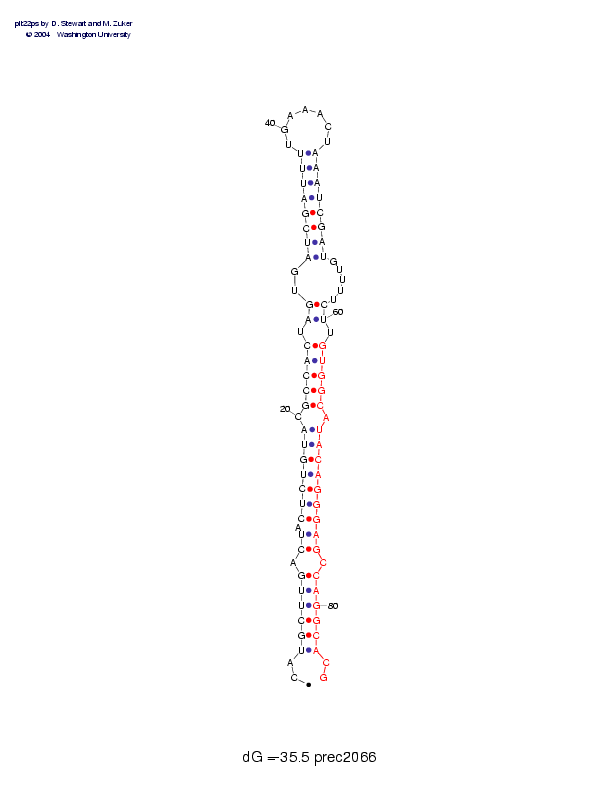
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
CAUGCUUGAC UACUCUGUAC GCCACUAGUG AUCGAUUUUG AAACUAAAUC GAUGUUUUCU 60
::((((((-( (-(((((((- (((((-((-- ((((((((__ _____))))) )))-----))
********* ********** *****
UGUGGCAUAC AGGGAGCCAG GCACG 85
-)))))-))) ))))))-))) )))::
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1495_132047-132132_63-86
- gnl|BL_ORD_ID|1495_132047+132132_1-24