| IGR sequence: | igr886#chr5#x1=3863724#l=4928 |
|---|---|
| Mature miRNA sequence: | UGUGUGCUCACUCUCUUCUGUCA |
| encoded miRNA: | MIR17e |
| Precursor location: | 3491 - 3586 (negative strand) |
| precursor length: | 96 (31 basepairs) |
| MIR position: | 74 - 96 (3491 - 3513) |
| MIR length: | 23 (21 paired bases) |
| miRNA location TIGR v3: | chr5:3867214<3867309 |
| miRNA location TIGR v5: | chr5:3867214<3867309 |
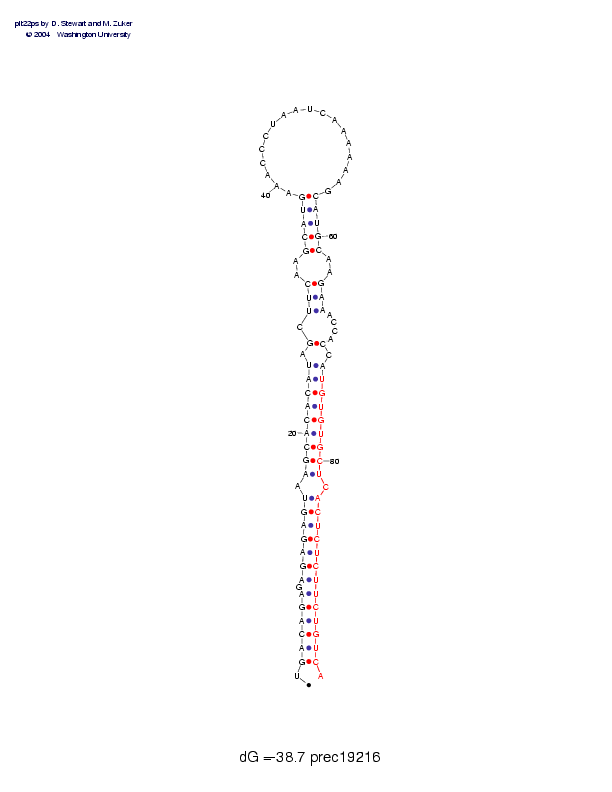
| Folding energy: | -38.67 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR156e |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
UGACAGAGAG AGAGUAAGCA CACAUAGCUU CAAGCAUGAA ACCCUAAUCA AAAAAGCAUG 60
:((((((-(( (((((-(((( (((((-(-(( (--(((((__ __________ ______))))
******* ********** ******
CAAGAAACCA CCAUGUGUGC UCACUCUCUU CUGUCA 96
)--)))---- )-)))))))) )-)))))))) ))))):
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|2024_146528-146637_88-110
- gnl|BL_ORD_ID|2024_146528+146637_1-23
- gnl|BL_ORD_ID|2021_81343+81452_1-23
- gnl|BL_ORD_ID|2021_81343-81452_88-110