| IGR sequence: | igr886#chr5#x1=3863724#l=4928 |
|---|---|
| Mature miRNA sequence: | GUGUGCUCACUCUCUUCUGUCA |
| encoded miRNA: | MIR17c |
| Precursor location: | 3491 - 3586 (negative strand) |
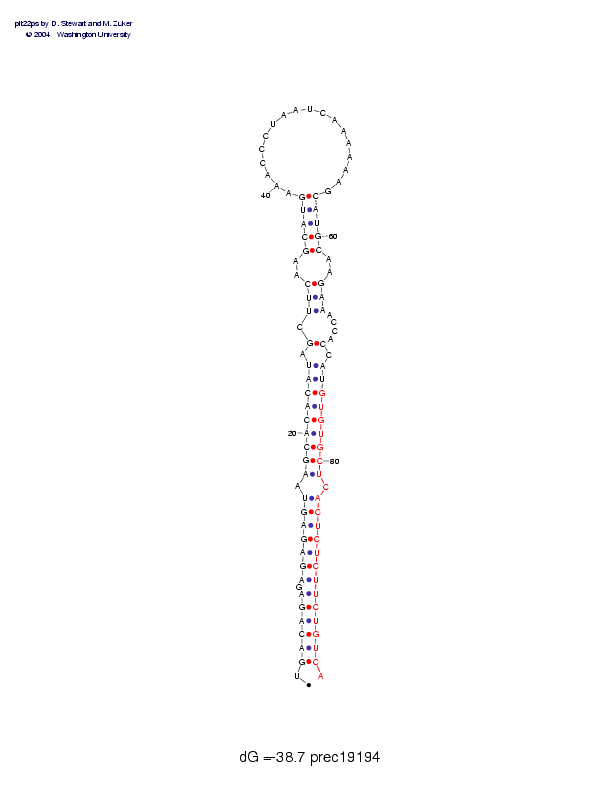
| precursor length: | 96 (31 basepairs) |
| MIR position: | 75 - 96 (3491 - 3512) |
| MIR length: | 22 (20 paired bases) |
| miRNA location TIGR v3: | chr5:3867214<3867309 |
| miRNA location TIGR v5: | chr5:3867214<3867309 |
| Folding energy: | -38.67 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR156e |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
UGACAGAGAG AGAGUAAGCA CACAUAGCUU CAAGCAUGAA ACCCUAAUCA AAAAAGCAUG 60
:((((((-(( (((((-(((( (((((-(-(( (--(((((__ __________ ______))))
****** ********** ******
CAAGAAACCA CCAUGUGUGC UCACUCUCUU CUGUCA 96
)--)))---- )-)))))))) )-)))))))) ))))):
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|2094_35724+35970_1-22
- gnl|BL_ORD_ID|2094_35724+35969_1-22
- gnl|BL_ORD_ID|2094_35724+35809_1-22
- gnl|BL_ORD_ID|2094_35724-35809_65-86
- gnl|BL_ORD_ID|2094_35724-35804_60-81
- gnl|BL_ORD_ID|342_115900-116035_115-136
- gnl|BL_ORD_ID|342_115900+115983_1-22
- gnl|BL_ORD_ID|342_115900-115983_63-84
- gnl|BL_ORD_ID|342_115900-115978_58-79
- gnl|BL_ORD_ID|62_1183-1268_65-86
- gnl|BL_ORD_ID|62_1183-1263_60-81
- gnl|BL_ORD_ID|62_1183+1268_1-22
- gnl|BL_ORD_ID|62_1183+1429_1-22
- gnl|BL_ORD_ID|62_1183+1428_1-22