| IGR sequence: | igr722#chr4#x1=3959983#l=2756 |
|---|---|
| Mature miRNA sequence: | UGCAUGAUAAGUUGUAAAGA |
| encoded miRNA: | MIR7 |
| Precursor location: | 2004 - 2212 (negative strand) |
| precursor length: | 209 (75 basepairs) |
| MIR position: | 190 - 209 (2004 - 2023) |
| MIR length: | 20 (15 paired bases) |
| miRNA location TIGR v3: | chr4:3961986<3962194 |
| miRNA location TIGR v5: | chr4:4996992<4997200 |
| Folding energy: | -55.70 |
| BLAST hit against RFAM Atha miRNAs: | none |
| Belongs to miRNAs-targets cluster: | none |
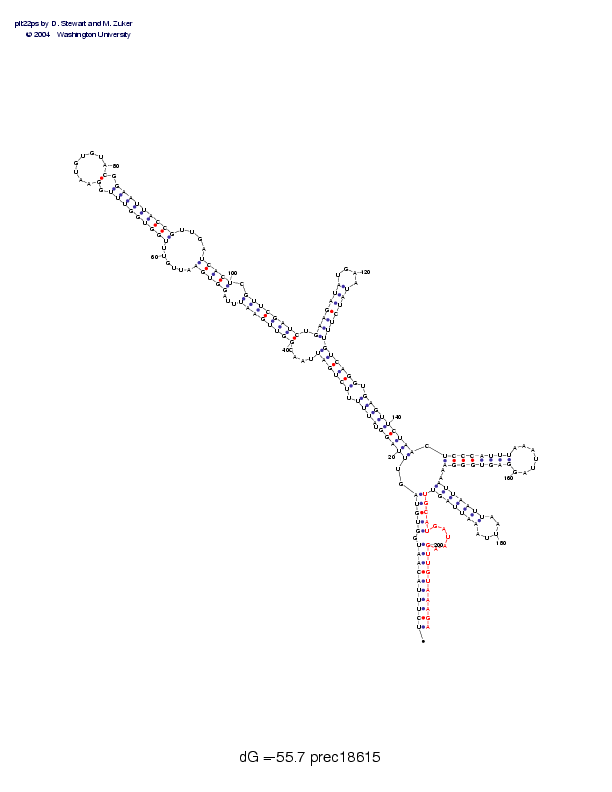
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
UCUUUACAAU GGUGUAGUUU AGGUAUUUUU CUGAUUAACG GUUGAAUUUA GGUGAAUUGU 60
(((((((((( -(((((,,<< <<<-<<<<-< <<<<<,,,,[ [[[[[[[--- [[[[[-----
UUGGUGGUUU GGAAUGUGUA CGGAAUUACC GUUGAUCACU CGUUCGAUCU GAAGAUAUGA 120
-[[[[[[[[[ -[________ ]-]]]]]]]] ]----]]]]] -]]]]]]]], [[[[[[[___
AUAUCUUUGU CAGGUGAGUU CUAACUCCCA UUUAAAUUAG GAGUGGGAAA AUUAAUUAAU 180
_]]]]]]]>> >>>>->>>>> >>>>,[[[[[ [[[_______ ]]]]]]]],, [[[[[[[___
* ********** *********
UAAAUUAGUU GCAUGAUAAG UUGUAAAGA 209
__]]]]]]]) ))))-----) )))))))))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1552_81436+81618_1-20