| IGR sequence: | igr648#chr5#x1=2838168#l=3068 |
|---|---|
| Mature miRNA sequence: | UCUCGGACCAGGCUUCAUUCC |
| encoded miRNA: | MIR22a |
| Precursor location: | 2464 - 2560 (positive strand) |
| precursor length: | 97 (33 basepairs) |
| MIR position: | 77 - 97 (2540 - 2560) |
| MIR length: | 21 (16 paired bases) |
| miRNA location TIGR v3: | chr5:2840631>2840727 |
| miRNA location TIGR v5: | chr5:2840631>2840727 |
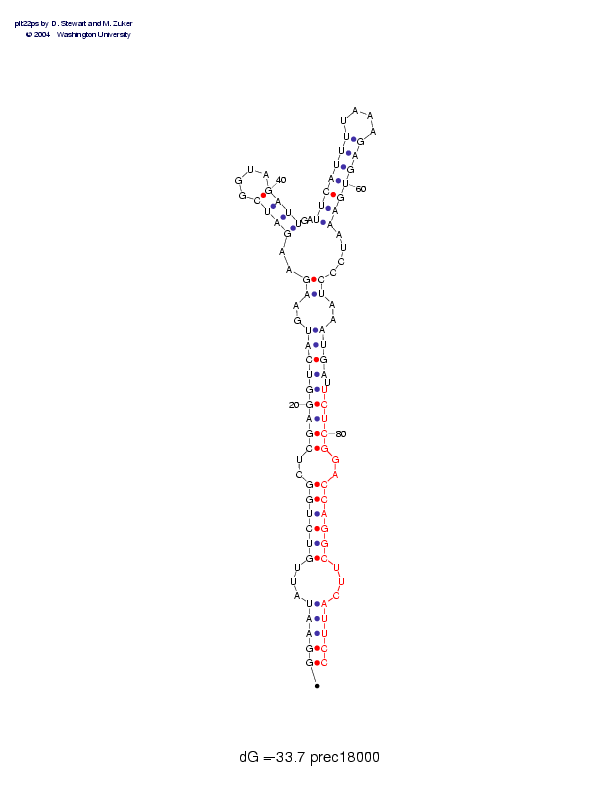
| Folding energy: | -33.70 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR166d |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
GGAAUAUUGU CUGGCUCGAG GUCAUGAAGA AGAUCGGUAG AUUGAUUCAU UUUAAAGAGU 60
(((((---(( ((((--(((( (((((--((, ,<<<<____> >>>,,<<<<< <<____>>>>
**** ********** *******
GAAAUCCCUA AAUGAUUCUC GGACCAGGCU UCAUUCC 97
>>>,,,,))- -))))-)))) )--))))))- --)))))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|2738_22961+23081_101-121
- gnl|BL_ORD_ID|2738_22961-23081_1-21
- gnl|BL_ORD_ID|2543_513-633_1-21
- gnl|BL_ORD_ID|2543_513+633_101-121