| IGR sequence: | igr5254#chr5#x1=25110924#l=3292 |
|---|---|
| Mature miRNA sequence: | CUCGGACCAGGCUUCAUUCCC |
| encoded miRNA: | MIR22 |
| Precursor location: | 2060 - 2146 (positive strand) |
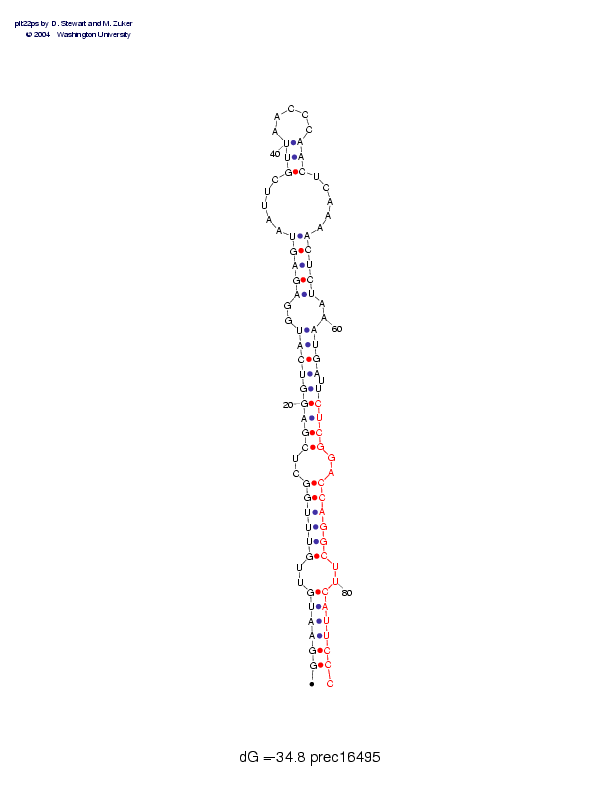
| precursor length: | 87 (29 basepairs) |
| MIR position: | 67 - 87 (2126 - 2146) |
| MIR length: | 21 (16 paired bases) |
| miRNA location TIGR v3: | chr5:25112983>25113069 |
| miRNA location TIGR v5: | chr5:25522041>25522127 |
| Folding energy: | -34.80 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR166g |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
GGAAUGUUGU UUGGCUCGAG GUCAUGGAGA GUAAUUCGUU AACCCAACUC AAAACUCUAA 60
((((((--(( ((((--(((( (((((--((( ((-----((( _____)))-- ---)))))--
**** ********** *******
AUGAUUCUCG GACCAGGCUU CAUUCCC 87
))))-))))) --))))))-- )))))):
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1999_118135+118264_110-130
- gnl|BL_ORD_ID|1300_66426+66555_110-130