| IGR sequence: | igr4997#chr3#x1=22923315#l=10188 |
|---|---|
| Mature miRNA sequence: | UCGGACCAGGCUUCAUUCCCC |
| encoded miRNA: | MIR70e |
| Precursor location: | 6887 - 6998 (positive strand) |
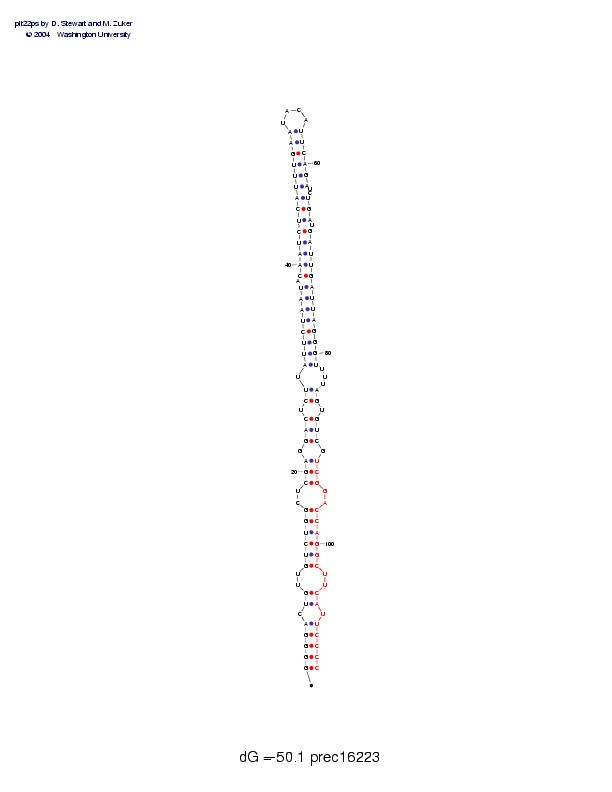
| precursor length: | 112 (43 basepairs) |
| MIR position: | 92 - 112 (6978 - 6998) |
| MIR length: | 21 (16 paired bases) |
| miRNA location TIGR v3: | chr3:22930201>22930312 |
| miRNA location TIGR v5: | chr3:22933185>22933296 |
| Folding energy: | -50.10 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR166b |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
GGGGACUGUU GUCUGGCUCG AGGACUCUUA UUCUAAUACA AUCUCAUUUG AAUACAUUCA 60
(((((-((-- ((((((--(( (-(((-((-( (((((((-(( (((((((((( ((____))))
********* ********** **
GAUCUGAUGA UUGAUUAGGG UUUUAGUGUC GUCGGACCAG GCUUCAUUCC CC 112
))--)))-)) )))))))))) )---))-))) -)))--)))) ))--))-))) ))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1466_100979-101071_1-21
- gnl|BL_ORD_ID|1466_100979+101071_73-93
- gnl|BL_ORD_ID|1425_155477+155569_73-93
- gnl|BL_ORD_ID|1425_155477-155569_1-21
- gnl|BL_ORD_ID|1343_7306+7372_47-67
- gnl|BL_ORD_ID|1343_7306-7372_1-21
- gnl|BL_ORD_ID|1337_41241-41307_1-21
- gnl|BL_ORD_ID|1337_41241+41307_47-67
- gnl|BL_ORD_ID|830_136567-136643_1-21
- gnl|BL_ORD_ID|830_136567+136643_57-77
- gnl|BL_ORD_ID|31_95866+95976_91-111