| IGR sequence: | igr4444#chr3#x1=20692045#l=9635 |
|---|---|
| Mature miRNA sequence: | GAUCAAUGCGAUCCCUUUGGA |
| encoded miRNA: | MIR20a |
| Precursor location: | 7608 - 7747 (negative strand) |
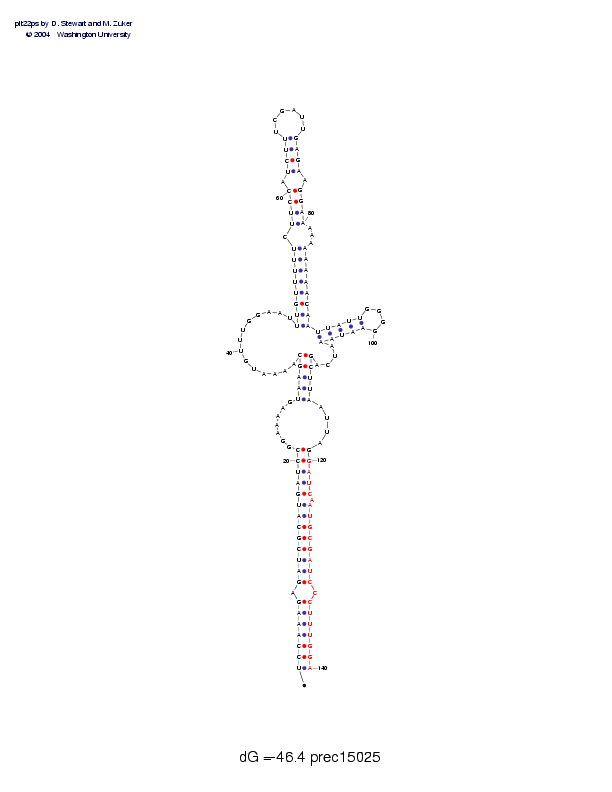
| precursor length: | 140 (46 basepairs) |
| MIR position: | 120 - 140 (7608 - 7628) |
| MIR length: | 21 (19 paired bases) |
| miRNA location TIGR v3: | chr3:20699652<20699791 |
| miRNA location TIGR v5: | chr3:20702636<20702775 |
| Folding energy: | -46.40 |
| BLAST hit against RFAM Atha miRNAs: | none |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
UCCAAAGAGA UCGCAUGAUC CGGAAAAGUA AGCAAAAUGU UUGGAAUUUG UUUUUCUUCC 60
(((((((-(( (((((((((( (-------(( (((,,,,,,, ,,,,,,,<<< <<<<<-<<<<
*
AUCUUUCGAU UGAGAAGGAA AAAAAAAACA AUUAUUGGGG AAUAAAUCAG CUUAAUUAGG 120
-<<<<_____ _>>>>->>>> --->>>>>>> ><<<<<____ >>>>>,,,,) ))))----))
********** **********
AUCAAUGCGA UCCCUUUGGA 140
)))-)))))) ))-)))))))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|2699_72476-72570_75-95
- gnl|BL_ORD_ID|2699_72476+72570_1-21
- gnl|BL_ORD_ID|2325_26142-26236_75-95
- gnl|BL_ORD_ID|2325_26142+26236_1-21