| IGR sequence: | igr4203#chr2#x1=19467319#l=3008 |
|---|---|
| Mature miRNA sequence: | GCACGUGCCCUGCUUCUCCA |
| encoded miRNA: | MIR83 |
| Precursor location: | 1909 - 1986 (negative strand) |
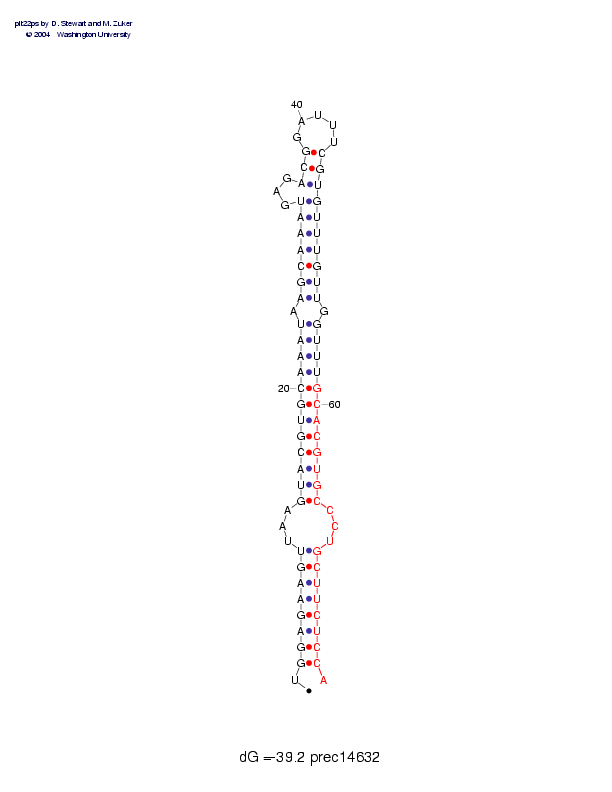
| precursor length: | 78 (30 basepairs) |
| MIR position: | 59 - 78 (1909 - 1928) |
| MIR length: | 20 (16 paired bases) |
| miRNA location TIGR v3: | chr2:19469227<19469304 |
| miRNA location TIGR v5: | chr2:19527840<19527917 |
| Folding energy: | -39.20 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR164a |
| Belongs to miRNAs-targets cluster: | cluster024 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
**
UGGAGAAGUU AAGUACGUGC AAAUAAGCAA AUGAGACGGA UUUCGUGUUU GUUGGUUUGC 60
:((((((((- --(((((((( ((((-((((( ((---(((__ ___))))))) )))-))))))
********** ********
ACGUGCCCUG CUUCUCCA 78
))))))---) ))))))):
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At1g56000.1 | 68408.m05886 amine oxidase-related | 1 | 1 |
Rice homologs
- gnl|BL_ORD_ID|396_77782-77855_55-74
- gnl|BL_ORD_ID|396_77782+77855_1-20