| IGR sequence: | igr4121#chr2#x1=19115407#l=13764 |
|---|---|
| Mature miRNA sequence: | CUUCGGACCAGGCUUCAUUCCCC |
| encoded miRNA: | MIR48 |
| Precursor location: | 9177 - 9312 (positive strand) |
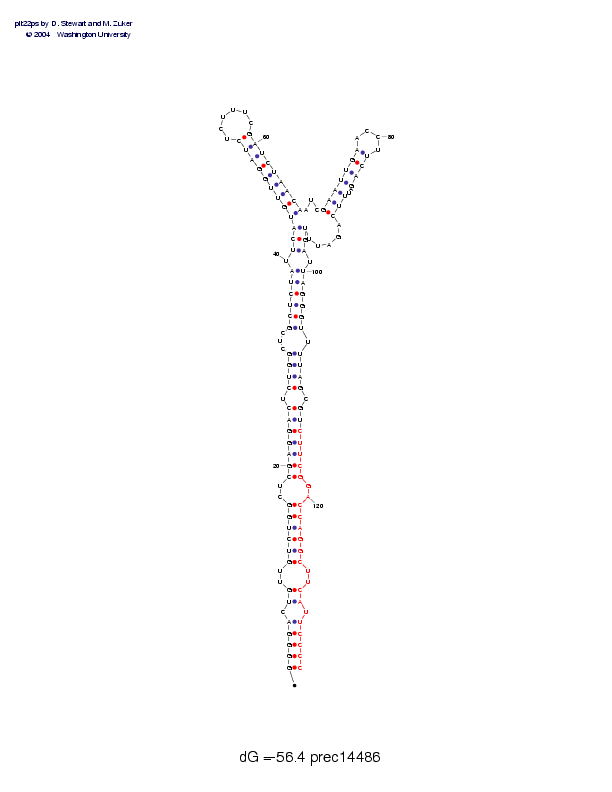
| precursor length: | 136 (49 basepairs) |
| MIR position: | 114 - 136 (9290 - 9312) |
| MIR length: | 23 (18 paired bases) |
| miRNA location TIGR v3: | chr2:19124583>19124718 |
| miRNA location TIGR v5: | chr2:19183196>19183331 |
| Folding energy: | -56.40 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR166a |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
GGGGACUGUU GUCUGGCUCG AGGACUCUGG CUCGCUCUAU UCAUGUUGGA UCUCUUUCGA 60
(((((-((-- ((((((--(( (((((-(((( ---((((((- (((<<<<<<< <<______>>
*******
UCUAACAAUC GAAUUGAACC UUCAGAUUUC AGAUUUGAUU AGGGUUUUAG CGUCUUCGGA 120
>>>>>>>,,, <<<<<<<___ _>>>>-->>> ,,,,,)))-) )))))-)))) -)))))))--
********** ******
CCAGGCUUCA UUCCCC 136
))))))--)) -)))))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1343_7306+7372_45-67
- gnl|BL_ORD_ID|1343_7306-7372_1-23
- gnl|BL_ORD_ID|1337_41243-41309_1-23
- gnl|BL_ORD_ID|1337_41243+41309_45-67