| IGR sequence: | igr3883#chr5#x1=19112826#l=3251 |
|---|---|
| Mature miRNA sequence: | UUAUAUUGUGAAACUGAGGGAGUAU |
| encoded miRNA: | MIR32d |
| Precursor location: | 1237 - 1457 (negative strand) |
| precursor length: | 221 (66 basepairs) |
| MIR position: | 197 - 221 (1237 - 1261) |
| MIR length: | 25 (24 paired bases) |
| miRNA location TIGR v3: | chr5:19114062<19114282 |
| miRNA location TIGR v5: | chr5:19523120<19523340 |
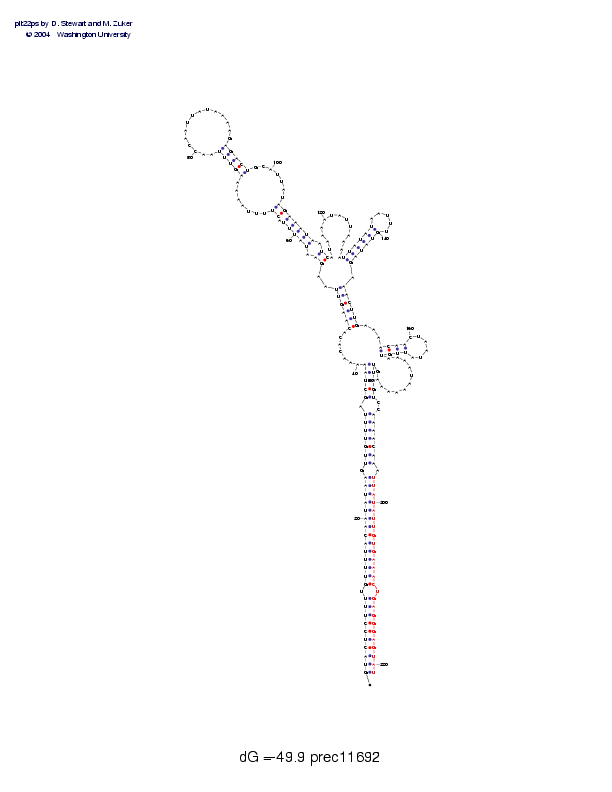
| Folding energy: | -49.90 |
| BLAST hit against RFAM Atha miRNAs: | none |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
GUACUCCUUU UGUUUUACAA UAUAAGUUGU UUUAGCUAAA AACACACAAG UUAAGAAUAU 60
(((((((((( -((((((((( (((((-(((( (((-(((((, ,,,,,,<<<< <<,,[[-[[[
UUACUUUUAA AAAGUUUAAC CAAUUAUAAA AGAGACUGCA UUAUAGAAAU AAUCAUAAAA 120
[[-[[----- --[[[[[___ __________ __]]]]]--- ----]]]]]] ]-]],,,,,,
UAUUAAAAUU AUAUAAUUUG UAUAGAAACU UGAAAACAAC UAAUAUUGUA AAAUAAAAAA 180
,,,,,,,,[[ [[[[_____] ]]]]],>>>> >>,,,[[[[_ _____]]]], ,,,,,,,,,,
**** ********** ********** *
GUUAGUCCAA AACAAAUUAU AUUGUGAAAC UGAGGGAGUA U 221
,)))))--)) )))))-)))) )))))))))) -))))))))) )
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1748_137894-138048_131-155
- gnl|BL_ORD_ID|1058_121799-121953_131-155
- gnl|BL_ORD_ID|1007_57541-57695_131-155
- gnl|BL_ORD_ID|3540_103742-104011_246-270
- gnl|BL_ORD_ID|3540_103742+104011_1-25
- gnl|BL_ORD_ID|3358_103086+103239_1-25
- gnl|BL_ORD_ID|3358_103086-103240_131-155
- gnl|BL_ORD_ID|3346_2534+2697_1-25
- gnl|BL_ORD_ID|3346_2534-2697_140-164
- gnl|BL_ORD_ID|3332_126630+126793_1-25
- gnl|BL_ORD_ID|3332_126630-126793_140-164
- gnl|BL_ORD_ID|3273_63773-63934_138-162
- gnl|BL_ORD_ID|3273_63773+63934_1-25
- gnl|BL_ORD_ID|3140_70689-70836_124-148
- gnl|BL_ORD_ID|3140_70689+70836_1-25
- gnl|BL_ORD_ID|2978_76491+76597_1-25
- gnl|BL_ORD_ID|2978_76491-76597_83-107
- gnl|BL_ORD_ID|2932_34800-34914_91-115
- gnl|BL_ORD_ID|2932_34800+34913_1-25
- gnl|BL_ORD_ID|2624_2307-2461_131-155
- gnl|BL_ORD_ID|2624_2307+2461_1-25
- gnl|BL_ORD_ID|2181_35612-35722_87-111
- gnl|BL_ORD_ID|2181_35612+35722_1-25
- gnl|BL_ORD_ID|1990_85042-85197_132-156
- gnl|BL_ORD_ID|1990_19707-19856_126-150
- gnl|BL_ORD_ID|1990_19707+19855_1-25
- gnl|BL_ORD_ID|1979_168201-168357_133-157
- gnl|BL_ORD_ID|1979_168201+168357_1-25
- gnl|BL_ORD_ID|1896_86720-86867_124-148
- gnl|BL_ORD_ID|1896_86720+86867_1-25
- gnl|BL_ORD_ID|1732_2831+2985_1-25
- gnl|BL_ORD_ID|1732_2831-2985_131-155
- gnl|BL_ORD_ID|1678_108214+108390_1-25
- gnl|BL_ORD_ID|1678_108214-108390_153-177
- gnl|BL_ORD_ID|1328_86253+86385_1-25
- gnl|BL_ORD_ID|1328_86253-86385_109-133
- gnl|BL_ORD_ID|1277_62469-62623_131-155
- gnl|BL_ORD_ID|1277_62469+62623_1-25
- gnl|BL_ORD_ID|1064_154011-154160_126-150
- gnl|BL_ORD_ID|1064_154011+154159_1-25
- gnl|BL_ORD_ID|1060_13287+13441_1-25
- gnl|BL_ORD_ID|1060_13287-13441_131-155
- gnl|BL_ORD_ID|915_42889+43050_1-25
- gnl|BL_ORD_ID|915_42889-43050_138-162
- gnl|BL_ORD_ID|849_135642-135827_162-186
- gnl|BL_ORD_ID|838_109893+110047_1-25
- gnl|BL_ORD_ID|838_109893-110047_131-155
- gnl|BL_ORD_ID|752_34660-34815_132-156
- gnl|BL_ORD_ID|715_26724-26878_131-155
- gnl|BL_ORD_ID|715_26724+26877_1-25
- gnl|BL_ORD_ID|357_67711-67865_131-155
- gnl|BL_ORD_ID|357_67711+67865_1-25
- gnl|BL_ORD_ID|120_89039-89194_132-156
- gnl|BL_ORD_ID|120_89039+89193_1-25
- gnl|BL_ORD_ID|24_91544+91698_1-25
- gnl|BL_ORD_ID|24_91544-91698_131-155