| IGR sequence: | igr3768#chr5#x1=18615936#l=2646 |
|---|---|
| Mature miRNA sequence: | GGCAUACAGGGAGCCAGGCAUA |
| encoded miRNA: | MIR40b |
| Precursor location: | 1331 - 1414 (positive strand) |
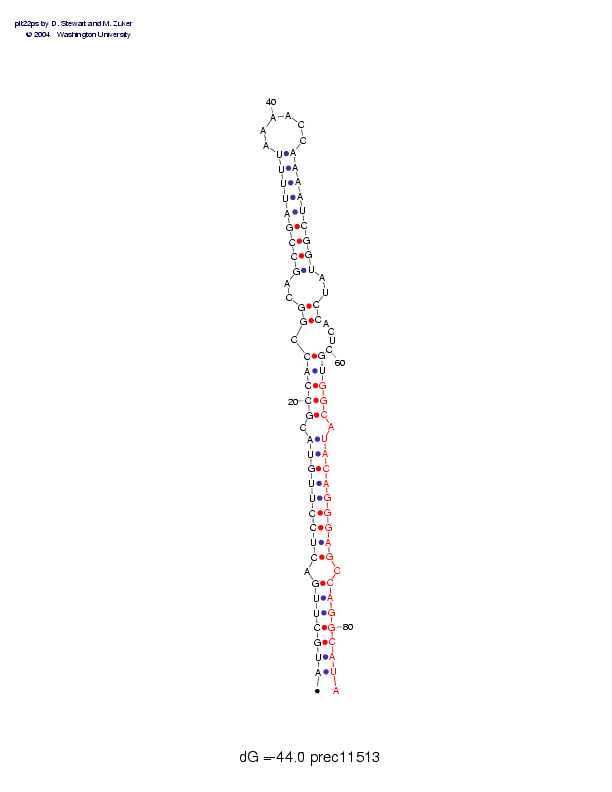
| precursor length: | 84 (32 basepairs) |
| MIR position: | 63 - 84 (1393 - 1414) |
| MIR length: | 22 (19 paired bases) |
| miRNA location TIGR v3: | chr5:18617266>18617349 |
| miRNA location TIGR v5: | chr5:19026324>19026407 |
| Folding energy: | -44.00 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR160c |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
AUGCUUGACU CCUUGUACGC CACCGGCAGC CGAUUUUAAA ACCAAAAUCG GUAUCCACUC 60
(((((((-(( (((((((-(( (((-((--(( (((((((___ ___))))))) ))--))----
******** ********** ****
GUGGCAUACA GGGAGCCAGG CAUA 84
)))))-)))) )))))-)))) ))):
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|3135_22620-22717_1-22
- gnl|BL_ORD_ID|2696_53306-53403_1-22
- gnl|BL_ORD_ID|255_18393-18490_1-22
- gnl|BL_ORD_ID|252_117310-117407_1-22