| IGR sequence: | igr3459#chr5#x1=17123821#l=3126 |
|---|---|
| Mature miRNA sequence: | UGAGGGGAAUGAAGCCUGGU |
| encoded miRNA: | MIR70b |
| Precursor location: | 654 - 750 (negative strand) |
| precursor length: | 97 (36 basepairs) |
| MIR position: | 1 - 20 (731 - 750) |
| MIR length: | 20 (15 paired bases) |
| miRNA location TIGR v3: | chr5:17124474<17124570 |
| miRNA location TIGR v5: | chr5:17533532<17533628 |
| Folding energy: | -39.10 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR166f |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** **********
UGAGGGGAAU GAAGCCUGGU CCGACAUCAU CAUCAUCAUC GUUAUAAUCA UGAGAUCAUG 60
::(((-(((( ((-((((((- -(((-((((( ((((((-((( -((((____) ))))))-)))
AGAUUGAAUG GUCUCGAGCC AGGCAUCAUU CACCUGA 97
)---))-))) ))-)))--)) ))))-))))) )-)))::
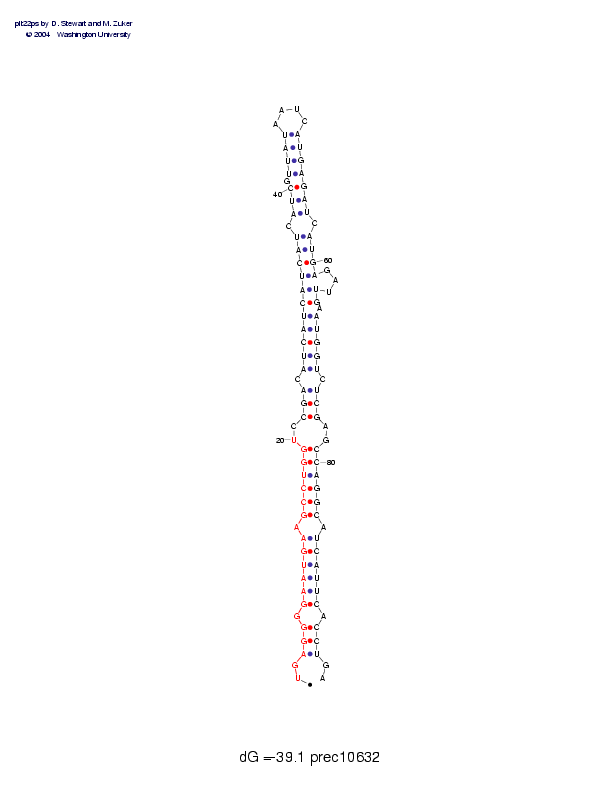
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|2976_134986+135110_106-125
- gnl|BL_ORD_ID|2976_134986-135110_1-20
- gnl|BL_ORD_ID|1743_126949+127073_106-125
- gnl|BL_ORD_ID|1743_126949-127073_1-20