| IGR sequence: | igr3459#chr5#x1=17123821#l=3126 |
|---|---|
| Mature miRNA sequence: | GGGAAUGAAGCCUGGUCCGA |
| encoded miRNA: | MIR6 |
| Precursor location: | 658 - 746 (negative strand) |
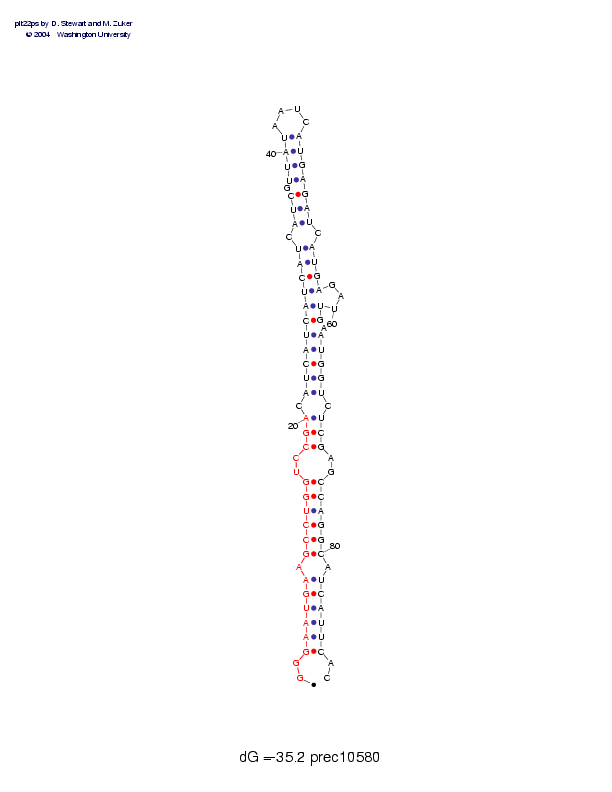
| precursor length: | 89 (33 basepairs) |
| MIR position: | 1 - 20 (727 - 746) |
| MIR length: | 20 (15 paired bases) |
| miRNA location TIGR v3: | chr5:17124478<17124566 |
| miRNA location TIGR v5: | chr5:17533536<17533624 |
| Folding energy: | -35.20 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR166f |
| Belongs to miRNAs-targets cluster: | cluster025 |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** **********
GGGAAUGAAG CCUGGUCCGA CAUCAUCAUC AUCAUCGUUA UAAUCAUGAG AUCAUGAGAU 60
::((((((-( (((((--((( -((((((((( ((-(((-((( (____))))) ))-))))---
UGAAUGGUCU CGAGCCAGGC AUCAUUCAC 89
))-)))))-) ))--)))))) -))))))::
Postscript file CT file PNG file fasta file Stockholm file
Targets:
| Nr. | Gene | Description | mismatches | miRNAs |
|---|---|---|---|---|
| 1 | At1g37050.1 | 68408.m04207 hypothetical protein | 2 | 1 |
Rice homologs
- gnl|BL_ORD_ID|1999_118134+118263_111-130
- gnl|BL_ORD_ID|1300_66425+66554_111-130