| IGR sequence: | igr3289#chr5#x1=16380038#l=5991 |
|---|---|
| Mature miRNA sequence: | GGGGAAUGAAGCCUGGUCCGA |
| encoded miRNA: | MIR70d |
| Precursor location: | 3657 - 3791 (positive strand) |
| precursor length: | 135 (38 basepairs) |
| MIR position: | 1 - 21 (3657 - 3677) |
| MIR length: | 21 (16 paired bases) |
| miRNA location TIGR v3: | chr5:16383694>16383828 |
| miRNA location TIGR v5: | chr5:16792752>16792886 |
| Folding energy: | -38.80 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR166e |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
********** ********** *
GGGGAAUGAA GCCUGGUCCG ACGUCAUUAA CCGUAAAAUG AUCAUUCAUG UAUAGACUAA 60
((((((((-- (-((((--(( (-(((-(((( (,,,,,,,,, ,,,<<<<<<_ _________>
UGAAUAAAGA AGAGACAUAU AUAUAUAAUC AAAUAUAGAU CUAAGUUAAG GGCCUCGUGC 120
>>>>>,,,,, ,,<<<--<<< <<________ __>>>>>--> >>,,)))))- )))-)))--)
CAGACAACAU UCCCC 135
)))-)--))) )))))
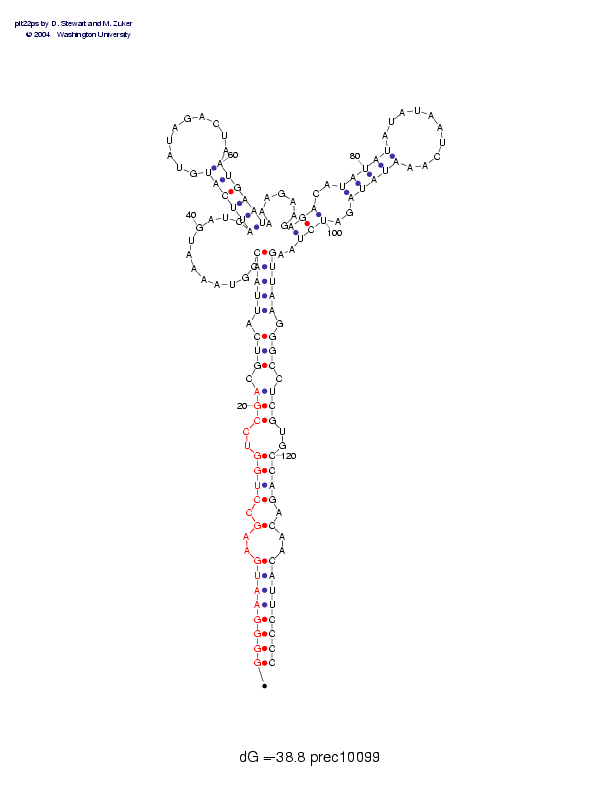
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1553_40801+40902_1-21
- gnl|BL_ORD_ID|1553_40801-40902_82-102
- gnl|BL_ORD_ID|1343_7352-7418_47-67
- gnl|BL_ORD_ID|1343_7352+7418_1-21
- gnl|BL_ORD_ID|1337_41287+41353_1-21
- gnl|BL_ORD_ID|1337_41287-41353_47-67