| IGR sequence: | igr3289#chr5#x1=16380038#l=5991 |
|---|---|
| Mature miRNA sequence: | UCGGACCAGGCUUCAUUCCCCUCAA |
| encoded miRNA: | MIR70a |
| Precursor location: | 3653 - 3795 (negative strand) |
| precursor length: | 143 (47 basepairs) |
| MIR position: | 119 - 143 (3653 - 3677) |
| MIR length: | 25 (20 paired bases) |
| miRNA location TIGR v3: | chr5:16383690<16383832 |
| miRNA location TIGR v5: | chr5:16792748<16792890 |
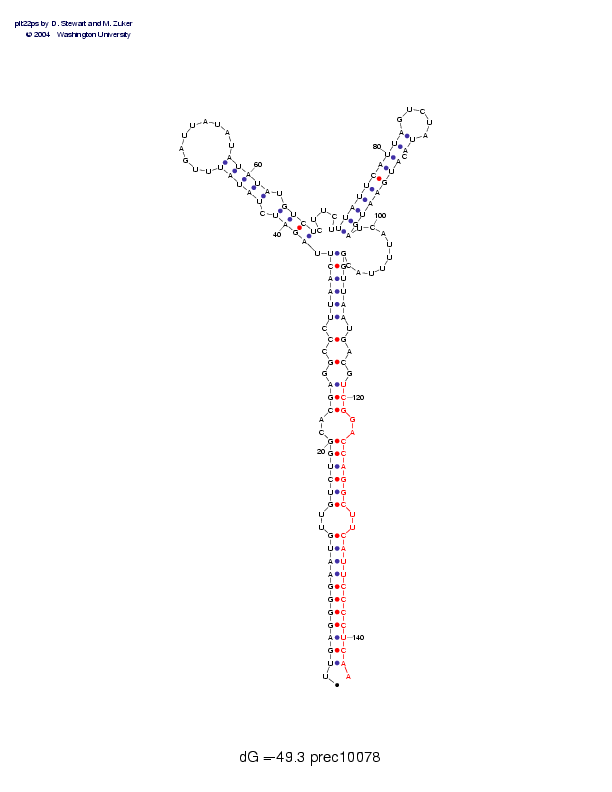
| Folding energy: | -49.30 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR166e |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
UUGAGGGGAA UGUUGUCUGG CACGAGGCCC UUAACUUAGA UCUAUAUUUG AUUAUAUAUA 60
:((((((((( ((--(((((( --(((-(-(- ((((((,<<< <-<<<<<___ _______>>>
**
UAUGUCUCUU CUUUAUUCAU UAGUCUAUAC AUGAAUGAUC AUUUUACGGU UAAUGACGUC 120
>>->>>>,,, ,,<<<<<<<< <<_____>>- >>>>>>>>,, ,,,,,,,))) )))-)-)-))
********** ********** ***
GGACCAGGCU UCAUUCCCCU CAA 143
)--))))))- -))))))))) )):
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|1466_101047-101147_77-101
- gnl|BL_ORD_ID|1425_155549-155649_77-101
- gnl|BL_ORD_ID|830_136619+136703_1-25
- gnl|BL_ORD_ID|830_136619-136703_61-85