| IGR sequence: | igr3268#chr5#x1=16266119#l=7505 |
|---|---|
| Mature miRNA sequence: | UGCUUGGACUGAAGGGAGCUCCCU |
| encoded miRNA: | MIR69 |
| Precursor location: | 2521 - 2692 (negative strand) |
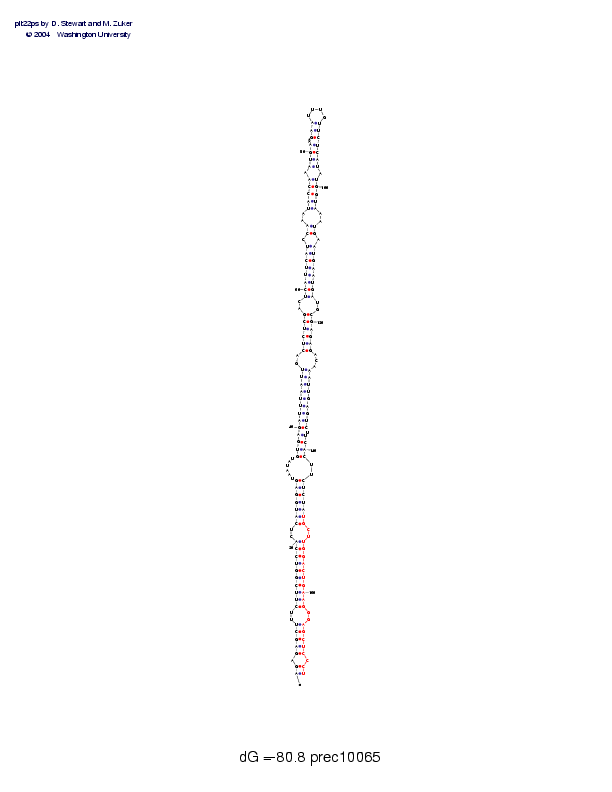
| precursor length: | 172 (65 basepairs) |
| MIR position: | 149 - 172 (2521 - 2544) |
| MIR length: | 24 (19 paired bases) |
| miRNA location TIGR v3: | chr5:16268639<16268810 |
| miRNA location TIGR v5: | chr5:16677697<16677868 |
| Folding energy: | -80.80 |
| BLAST hit against RFAM Atha miRNAs: | ath-MIR319b |
| Belongs to miRNAs-targets cluster: | none |
Sequence and secondary structure:
Presumed mature miRNA positions are indicated by *'s.
AGAGAGCUUU CUUCGGUCCA CUCAUGGAGU AAUAUGUGAG AUUUAAUUGA CUCUCGACUC 60
((-(((((-- (((((((((( --(((((((- -----((((( ((((((((-- ((((((--((
AUUCAUCCAA AUACCAAAUG AAAGAAUUUG UUCUCAUAUG GUAAAUGAAU GAAUGAUGCG 120
((((((-((- -(((((-((( (--(((____ )))))))-)) )))--))-)) ))))))--))
** ********** ********** **
AGAGACAAAU UGAGUCUUCA CUUCUCUAUG CUUGGACUGA AGGGAGCUCC CU 172
))))---))) ))))))-))) )--))))))) --)))))))) ))--)))))- ))
Postscript file CT file PNG file fasta file Stockholm file
Targets:
No Targets found for this miRNA
Rice homologs
- gnl|BL_ORD_ID|493_28731+28908_1-24
- gnl|BL_ORD_ID|493_28731-28908_155-178