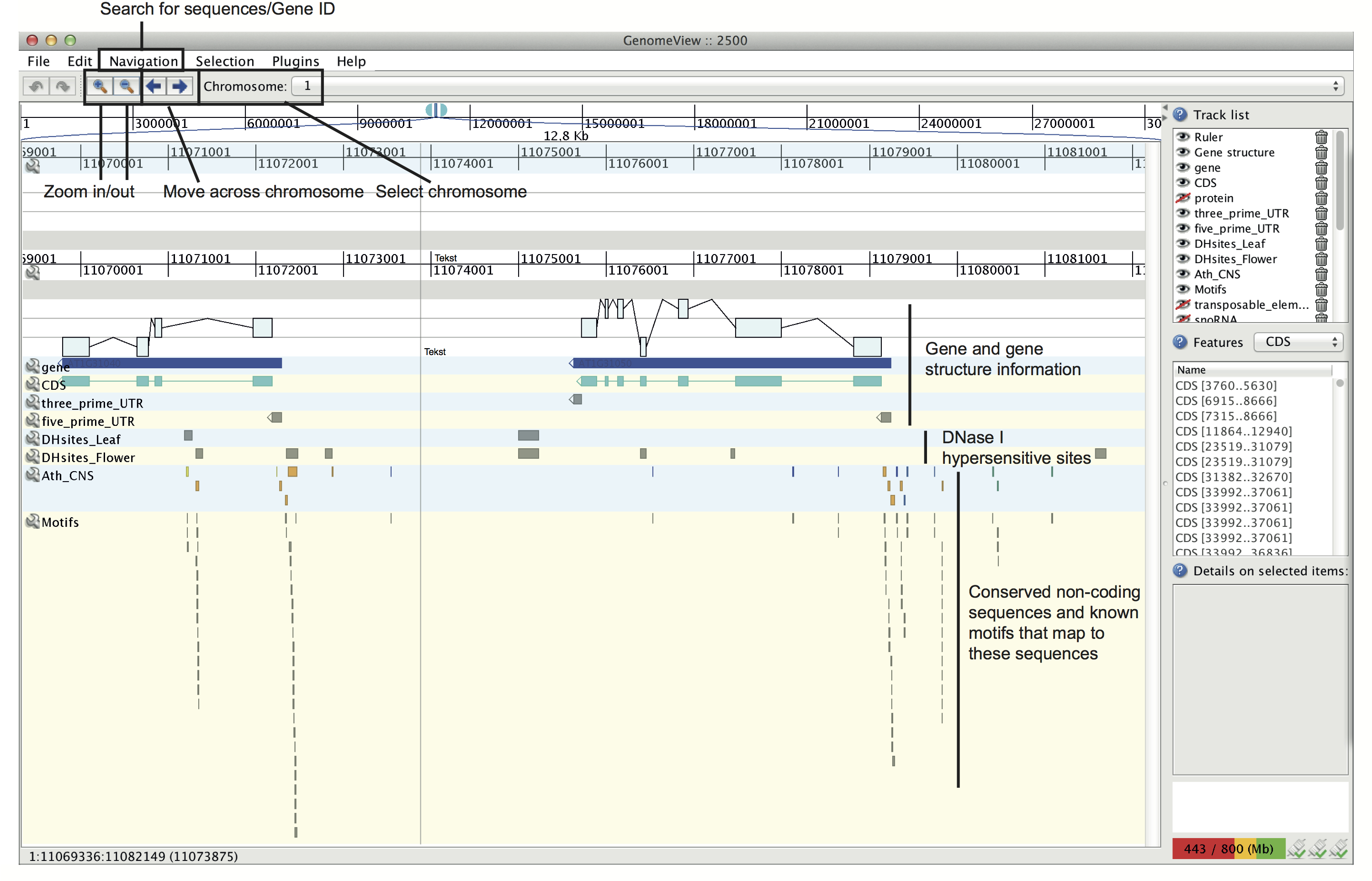
Inference of transcriptional networks in Arabidopsis thaliana through conserved non-coding sequence analysisJan Van de Velde*, Ken S Heyndrickx* and Klaas Vandepoele*Contributed equally Corresponding author:AbstractTranscriptional regulation plays an important role in establishing gene expression profiles during development or in response to (a)biotic stimuli. Transcription factor binding sites (TFBS) are the functional elements that determine transcriptional activity and the identification of individual TFBS in genome sequences is a major goal to inferring regulatory networks. We have developed a phylogenetic footprinting approach for the identification of conserved non-coding sequences (CNSs) across 12 dicot plants. Whereas both alignment and non-alignment-based techniques were applied to identify functional motifs in a multi-species context, our method accounts for incomplete motif conservation as well as high sequence divergence between related species. We identified 69,361 footprints associated with 17,895 genes. Through the integration of known TFBS obtained from literature and experimental studies, we used the CNSs to compile a gene regulatory network containing 40,758 interactions, of which two-thirds act through binding events located in DNase I hypersensitive sites. This network shows significant enrichment towards in vivo targets of known regulators and its overall quality was confirmed using five different biological validation metrics. Finally, through the integration of detailed expression and function information, we demonstrate how static CNSs can be converted into condition-dependent regulatory networks, offering new opportunities for regulatory gene annotation. Reference: Inference of Transcriptional Networks in Arabidopsis through Conserved Noncoding Sequence Analysis. Van de Velde, J., Heyndrickx, K.S., and Vandepoele, K. Plant Cell, 2014. 26(7): p. 2729-45 Supplementary DataA quick overview of the different tracks and features in the genome browser are displayed below. An extended Genomeview manual can be found here. |