Gene Family tutorial
This tutorial will be of use mainly for experimental biologists that have their favorite gene(s) and want to study the organization and evolution of their corresponding gene families. Using the Search bar & BLAST interface you can find your favorite gene(s) or related homologs from other species. Once you've found the correct gene family, there are functions to find sub-families, orthologs, paralogs, protein domains, phylogenetic trees, etc.
This tutorial will show how to use the Similarity heatmap and Synteny Plot. Also it will explain how to start jalview to inspect multiple sequence alignments and how the precomputed trees can be viewed with the tree explorer.
Gene Family Page
Once an interesting gene is found (here we'll use gene AT2G17380 as an example), the corresponding gene page provides detailed information about the raw annotation (eg. intron-exon structure, functional annotation through InterPro or GO) as well as more advanced features (eg. duplication type, homologous gene families). To explore the different properties of a gene family, this protocol can be used:
- At the gene page click the gene family ID to go to the gene family page (Result). Note that the next steps can also be used to study the sub family.
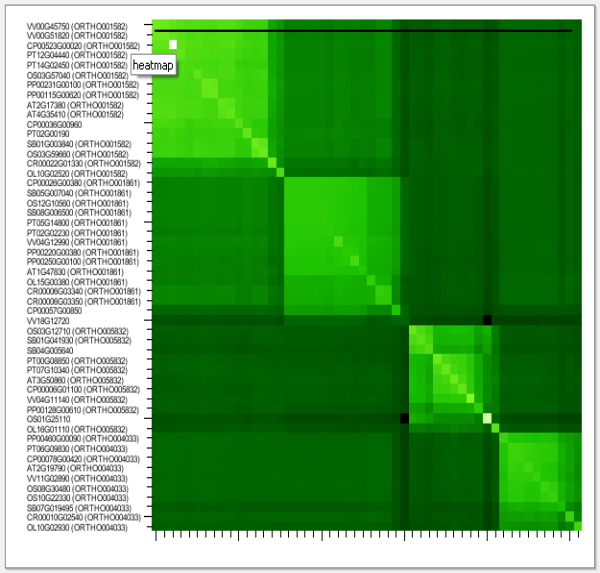
- Here a pie-chart (requires Flash) is provided that shows the distribution of the genes in the family over the different species present in PLAZA. This representation can give a first indication whether the family is expanded or lost in certain species.
- In the toolbox click on the Toolbox→View the similarity heatmap of this gene family. This will show the Similarity heatmap (Figure 1). Here several sub-families can be identified by inspecting the BLAST similarity scores. Clicking a gene ID will redirect you to the corresponding gene page, whereas clicking the heatmap will redirect you to the corresponding sub family (if available). Result
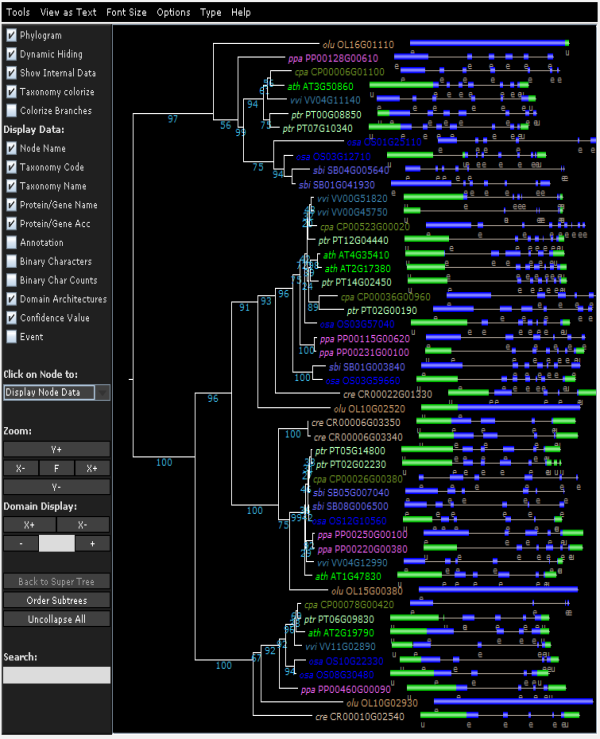
- Head back to the gene family page and inspect the phylogenetic tree : Toolbox→Explore the phylogenetic trees of this gene family. (requires the Java Runtime Environment) Result By default the domains are shown in the tree.
- In the tree view try clicking View phylogenetic tree with associated gene structure information. This will show the intron-exon structure of all the genes in the tree (Figure 2).
- When View basic phylogenetic tree showing speciation/duplication events nodes will be color coded based on the tree reconciliation. (Green = speciation, Red = duplication)
- By analyzing the phylogenetic tree paralogs (see above), true orthologs and different sub-types (or out-paralogs) can be identified. Note that apart from the protein domains also the gene structure and speciation/duplication events can be visualized.
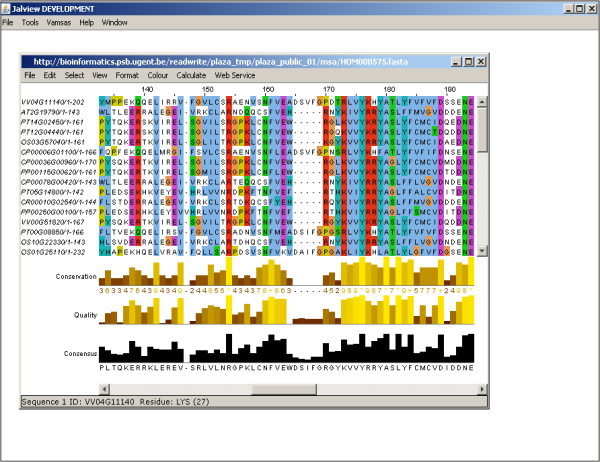
- Apart from the phylogenetic tree, also the multiple sequence alignment can be viewed. At the gene family page in the Toolbox click View the multiple sequence alignement of this gene family. to start Jalview (webstart). This alignment editor generates an alignment view that can be used to detect highly conserved regions or protein motifs (Figure 3).
- A complementary tool is the WGMapping tool, that can be used to view the distribution of a certain gene family across a genome. At a gene family page Toolbox→View the genome wide organization of this gene family.
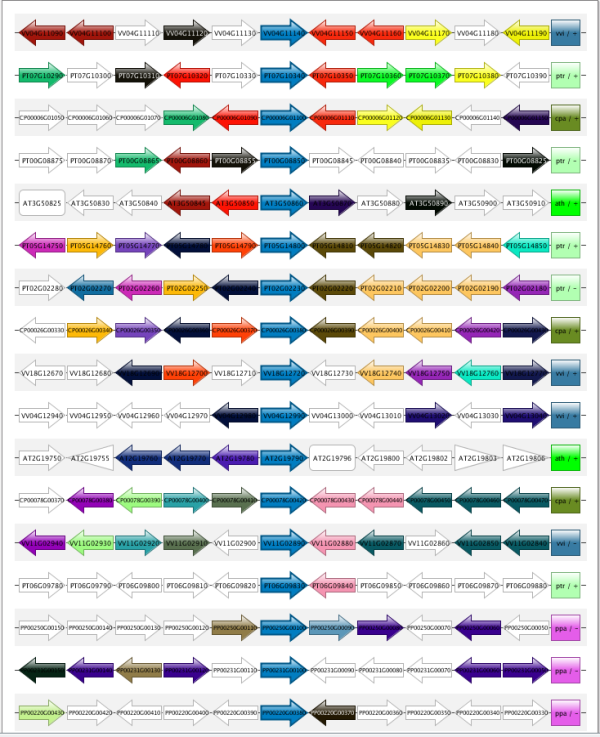
- Also a Synteny Plot has been created to track down positional orthologs. To get to this view goto : Toolbox→Explore the local gene organization for homologous genes (Figure 4).
Other
The Similarity heatmap, Synteny plot and WGMapping can also be reached through the Analyze button in the menu.
For those that paid close attention, there is one more option in the toolbox : View the functional annotation associated with this gene family. this function will be explained in another part of the tutorial about functional annotation. The WGDMapping tool can be used to map other features as well in case it's started from the menu.