Functional annotation
In the introduction we already mentioned that GO terms and InterPro domains can be used to search genes in PLAZA. However, there are several other pages that allow users to browse and analyze functional annotation.
Parent-child GO term relationships
In this section we will explain the basic functionality of PLAZA when working with GO terms. Here we will demonstrate how to find genes associated with a certain GO term and explain how the relationships between GO terms can be used to retrieve more detailed or more general information.
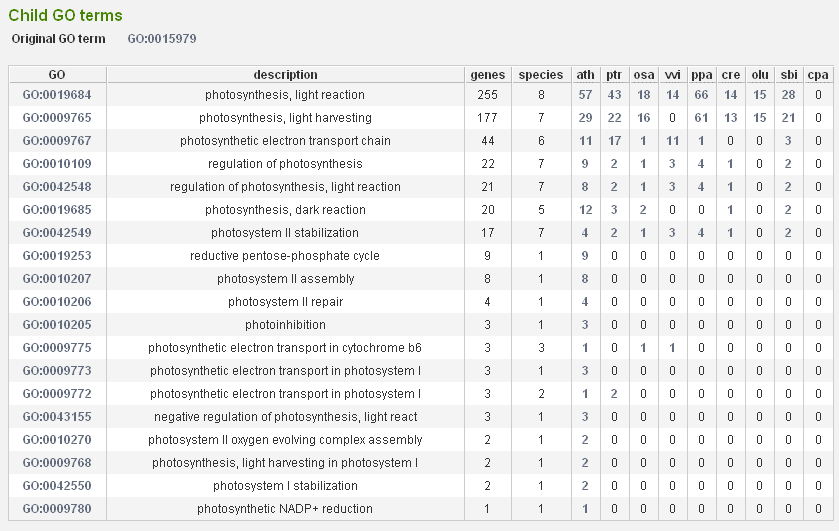
- First start with finding the GO term for photosynthesis using the search function. (Result)
- Already from the pie chart on the GO page it's clear that all species have genes associated with this label. Note that the green algae Ostreococcus and Chlamydomonas have less genes.
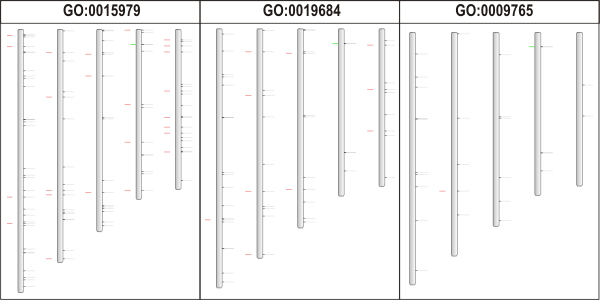
- We select Toolbox→View the genome wide organization of this GO term. By selecting Arabidopsis thaliana as organism, we can now see where the genes involved in photosynthesis are located on the chromsomes in Arabidopsis thaliana. All red ticks represent block duplicated genes covering ~28% of all Arabidopsis genes annotated using the GO term GO:0015979 Photosynthesis.
- On the GO page we can click in the toolbox on Toolbox→View child GO terms. (Result) Note that the more specific GO terms typically contain less genes since they represent more specific or detailed biological information (typically obtained through an experimental approach). Although by default the most specific GO term is associated to a gene product, the parent-child relationships between GO terms make it possible to search for all genes related to a more general GO term in PLAZA (i.e. a gene annotated with the GO term photoinhibition is by default also annotated with the term photosynthesis).
- Here we select photosynthesis, light reaction GO:0019684, and again we view the genome wide organization of this GO term. Repeat this operation with child GO term photosynthesis, light harvesting GO:0009765.
- Finally we end up with 3 images depicting the genome wide organization of genes associated with GO terms that are connected through direct parent-child relationships in the GO graph.
Mapping InterPro domains on gene families and phylogenetic trees
Here we will demonstrate how to explore InterPro domains mapped on a set of gene families and the corresponding phylogenetic tree(s).
- First we start by searching for the keyword auxin in the toolbar (using GO/InterPro description).
- We select the IPR003676 InterPro domain, which has Auxin responsive SAUR protein as description.
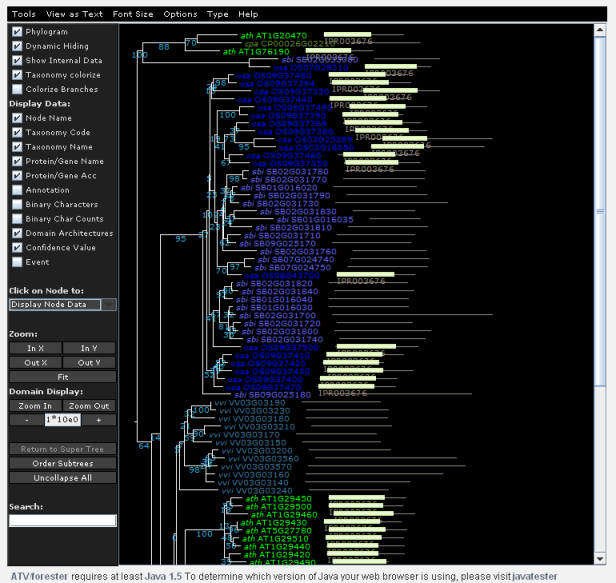
- In the toolbox, we click on Toolbox→View the associated gene families. (Result) This table shows all the gene families covering genes annotated with this InterPro domain.
- Here we can use the ATV tree viewer 'Explore the phylogenetic trees of this gene family' to further analyze the distribution of this InterPro domain in this gene family.
Browsing genes in species lacking functional annotation
While PLAZA contains a functional annotation (~free-text gene description or InterPro/GO associations) for most organisms, some species lack GO or InterPro annotations. In this tutorial we will once again use the photosynthesis example in order to discover genes in Carica Papaya which, based on sequence similarity, are associated with photosynthesis in plants.
- We start with finding the GO term for photosynthesis using the search function (Result)
- We arrive at the GO page and in the toolbox on this page, click on Toolbox→View the associated gene families. (Result)
- The table on this page displays which gene families contain genes associated with a certain GO term (in this case GO:0015979), and other information is also given, eg. How many genes are there in total in the gene families?, How many of these genes are annotated using this GO term?, etc.
- When we consider it likely that most green plants will have a rather similar amount of genes related to photosynthesis, the first gene family (HOM000102) in the table seems a likely candidate for further investigation (~ two thirds of all genes are annotated with this GO term).(Result)
- The gene family page shows that Carica Papaya has 17 genes in this gene family. Clicking on the pie-chart part of Carica Papaya redirects us to the list of Papaya genes in gene family HOM0000102 (Result)
- For each of these genes, we have no direct functional information. However, by clicking on a gene we are redirected to its respective gene page. At each gene page, we can BLAST this gene against the NCBI database of genes by clicking on Toolbox→View BLAST hits against NCBI's protein database.
- When examining the BLAST results for gene CP00008G00050 it is quite apparent that most of the BLAST hits have a description related to chlorophyll binding and can thus be said to be associated with photosynthesis (with a highly significant e-value). Therefore, the associated gene families provide a practical entry point to transfer functional annotations between species using gene homology.