Step 1:
OPTION 1: one or more query genes
STEP 1 Help (Double click to close)
By default the input data are GRMZM codes of your genes of interest. Other maize identifiers, such as PLAZA v2.0 identifiers (e.g. ZM08G15930),
Affymetrix probe identifiers (e.g. ZM.17362.s1_at), or Maize Oligonucleotide Array identifiers (e.g. MZ00002380 from Arizona Maize Array) are also
accepted as input. Additionally, the program allows also orthologous gene identifiers from Arabidopsis using the AGI code (TAIR identifiers, e.g. At2g33610),
orthologous gene locus identifiers from rice (TIGR identifiers without the LOC_ prefix, e.g. Os02g10060).
Note that, if an orthologous gene (from Arabidopsis and/ or Oryza) is given and if it is part of a group of orthologous genes with a many-to-many relationship (extracted using
OrthoMCL results from PLAZA
v2 framework)
then from this orthologous group all maize genes will taken into account.
|
|
- Paste a list of GRMZM code in the text area on the left
- The GRMZM code may be seperated by tabs, spaces, semicolon, commas and newlines
- The GRMZM code are case insensitive allowing to mingle upper and lower case letters, e.g. GrMzM2G064328,
gRMzm2g121510, or
GRMZM2g007734
- Homologous genes from rice or Arabidopsis as well as other maize gene identifiers are also allowed as input, e.g. At4g04885,
Os02g10060, or
ZM08G15930 ?
|
OPTION 2: upload a file containing a list of gene pairs
Step 3: Click for guidelines
Set other options: ?
STEP 3 Help (Double click to close)
Multiple expression datasets:
Expression correlation can be measured based on multiple expression datasets (selected in Step 2) simultaneously. Only the Pearson correlation coefficient is available in this option.
Pairwise comparisons:
All possible combinations between query genes are made. A correlation coefficient is calculated for each pair of genes
Neighbours:
Add extra genes. Every query gene is compared to the complete Arabidopsis genome. A gene pair with a correlation coefficient above the chosen thresholds is reported.
Thresholds:
Two thresholds can be chosen.
- Correlation coefficients higher and lower than a certain value (if given) are reported.
- The top X genes with highest correlation coefficients are reported, or bottom genes when comparing negative correlation coefficients. Genes with same correlation coefficients are grouped
The most limiting threshold always has priority.
(e.g. If you ask for top 10 neighbours but only 6 meet the first threshold's requirement, only 6 are shown. If you ask for top 10 neighbours but many more meet the first threshold's requirement, only 10 are shown.)
Relations between neighbours:
If you want to test if the neighbours of your query gene are co-expressed, you need to choose this option.
Cytoscape webstart Help (Double click to close)
Cytoscape will launch through JAVA™ Web Start Launcher
You may need to update JAVA™ for the Cytoscape Web Start.
Get the latest JAVA™ Runtime Environment
You can also download and install the latest version of Cytoscape locally on your computer.
Visualization legend:
| Correlation networks |
| Edge color | 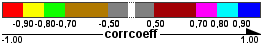 |
| Query gene |  |
| Protein-protein interactions |
| Edge color | black |
| Edge width | the more references, the wider the edge |
| Edge style | = experimental
= predicted |
| Query gene |  |
| COR and PPI |
| Query gene |  |
 Localization pie colors Localization pie colors
(the size of a pie slice represents how much
a value is referenced compared to the others) |
| | CYTOSKELETON |
| | PEROXISOME |
| | OTHER |
| | CELLPLATE |
| | NUCLEOLUS |
| | CYTOSOL |
| | CHLOROPLAST |
| | NUCLEUS |
| | MITOCHONDRIA |
| | ENDOSOME |
| | PLASTID |
| | VACUOLE |
| | ENDOPLASMICRETICULUM |
| | EXTRACELLULAR |
| | PLASMAMEMBRANE |
| | GOLGI |
Guidelines (Double click to close)
- Start with stringent parameters depending on the number of query genes:
- High correlation coefficient (e.g. 0.8)
- Low number of neighbours (e.g. 10)
- Gradually loosen the stringency of the parameters
- When the co-expression network is too large, a warning will be given. A text output instead of a visual Cytoscape output can be chosen
- When selecting multiple expression datasets, the number of co-expression links can rapidly increase as different expression dataset can yield complementary results (particularly for atleast=1).
|
Step 2:
STEP 2 Help (Double click to close)
This Cornet version contains different types of expression compendiums performed by GEO
Single platform compendium (absolute values)
- Affymetrix Maize Genome Array Compendium including 128 experiments (GEO, 12/2010).
- Nimblegen Maize Whole-Genome Microarray 385K (VersionV1_4a.53) taken from Sekhon et
al., (2011) including 60 experiments (GEO, 06/2011).
STEP 2 Help (Double click to close)
This Cornet version contains different types of expression compendiums performed by GEO
Single platform compendium (absolute values)
- Affymetrix Maize Genome Array Compendium including 128 experiments (GEO, 12/2010).
- Nimblegen Maize Whole-Genome Microarray 385K (VersionV1_4a.53) taken from Sekhon et
al., (2011) including 60 experiments (GEO, 06/2011).
Single platform compendiums based on comparison to reference experiments (ratios)
- ISU Maize 12k cDNA Generation II (144 exps, 4472 genes, ratios, 02/2010) contains
global gene expression profiling of developing maize anthers
- Agilent Maize 21K v1.0 Array (24 exps, 9001 genes, ratios, 02/2010) contains
comparative transcriptome profiling of maize lines
- Agilent Maize 22K v2.0 Array (36 exps, 9907 genes, ratios, 02/2010) contains
transcriptome profiling of maize anthers using genetic ablation
- ZmDB 606-Immature Ear Microarray (37 exps, 1155 genes, ratios, 02/2010) contains UV
radiation gene profiling and expression non-additivity in immature ears of a heterotic F1 maize
- Maize 12K cDNA Generation II (36 exps, 4512 genes, ratios, 02/2010) contains
experiments related to vegetative phase change
Combined platform compendiums based on comparison to reference experiments (ratios)
- Maize Oligo Compendium I (GPL7209 and GPL6460) (26 exps, 14594 genes, ratios, 02/2010) contains experiments related to
comparison of RNAi chc101 and mdb101 transgenic plants as well as maize anthers and pollen ontogeny
- Maize Oligo Compendium II(GPL1990_1991, GPL1992_199, and GPL5439_5440)(80 exps, 15583 genes, ratios, 02/2010) contains experiments related to gene expression profiling
of bundle sheath and mesophyll cell types, maize root, aluminum toxicity and tolerance responces, maize leaf at heading stage, kernel development, de-regulation of maize C4
photosynthetic development in a mesophyll cell
- Maize SAM Array Compendium (1.0, 2.0, and 3.0 Array
)(36 exps, 10089 genes, ratios, 02/2010) contains experiments related to global analysis in shoot apical meristem and histologiocal layers of the shoot as well as transcriptome
analyses of narrow sheat shoot apical meristem and of ragged seedling shoot apical meristem
Select MicroArray data: ?
Note, it will use all genes where the selection criteria are valid for the
in at least
selected datasets:
Affymetrix Maize Genome Array Compendium
(128 exps, 9846 genes, absolute values, 12/2010)
Nimblegen Maize Whole-Genome Microarray 385K (VersionV1_4a.53)
(60 exps, 22600 genes, absolute values, 06/2011)
|
