Browsing conserved TF binding sites tutorial
These tutorials will give a basic insight into how to browse and interpret conserved TF binding sites. By starting from either a target gene, a TF (transcription factor) gene, or a known binding site, the user can easily find the other associated information he is looking for.
For a TF of interest, identify target genes with a conserved binding site
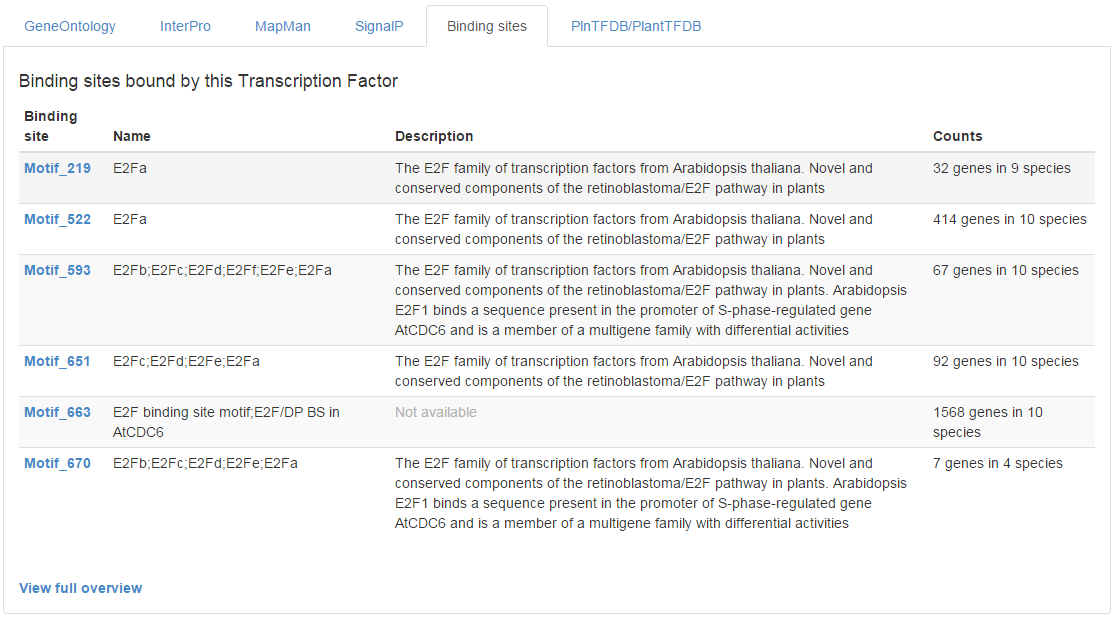
- On each gene page encoding a TF, the tab ‘Binding sites’ reports the associated binding site information (shortly Motifs) (see Figure 1).
- For each motif, the Binding site page lists the associated TF, a short description and literature info, the sequence logo and the number of associated genes with a conserved binding site (see Figure 2).
- Two pie charts report the breakdown of the number of target genes per species and per sequence location (see Figure 2).
- The Toolbox offers additional analysis options (see section ‘Functional analysis’).
- Associated TFs lists related TFs with similar motifs.
- Gene list offers customizable Download option.
For a gene of interest, identify the conserved binding sites
- For each gene the Gene page now offers an extra option in the Toolbox, ‘Explore ...the conserved binding sites (upstream/downstream,intron)’
- This options allows browsing conserved motifs, the associated TFs as well as the location and position of the conserved motifs (see Figure 3).
Functional analysis
In the Toolbox, different options are available to analyse the target genes of a specific TF in more detail.
- 'View the associated gene families' offers an overview of the target genes with a conserved TF binding sites summarized per gene family. Based on the example for Motif_522, it is clear that the 414 target genes belong to 107 gene families (see Figure 4).
- By sorting the table using header ‘#associated genes’, it immediately becomes clear that three families with the largest number of target genes are linked with DNA replication and repair (hoover over gene family HOM identifiers to see Functional annotation).
- Similarly, using the option ‘Explore the associated GO or InterPro annotation of this binding site’, functional annotation of the target genes can be obtained.
- To explore similar binding sites, the option ‘View ... the binding sites with a similar sequence.’ makes it possible to identify similar TF binding sites. The ‘Minimum similarity percentage’ options allows the selection of more divergent (dissimilar) binding sites.
Detailed characterization TF target genes using the PLAZA Workbench
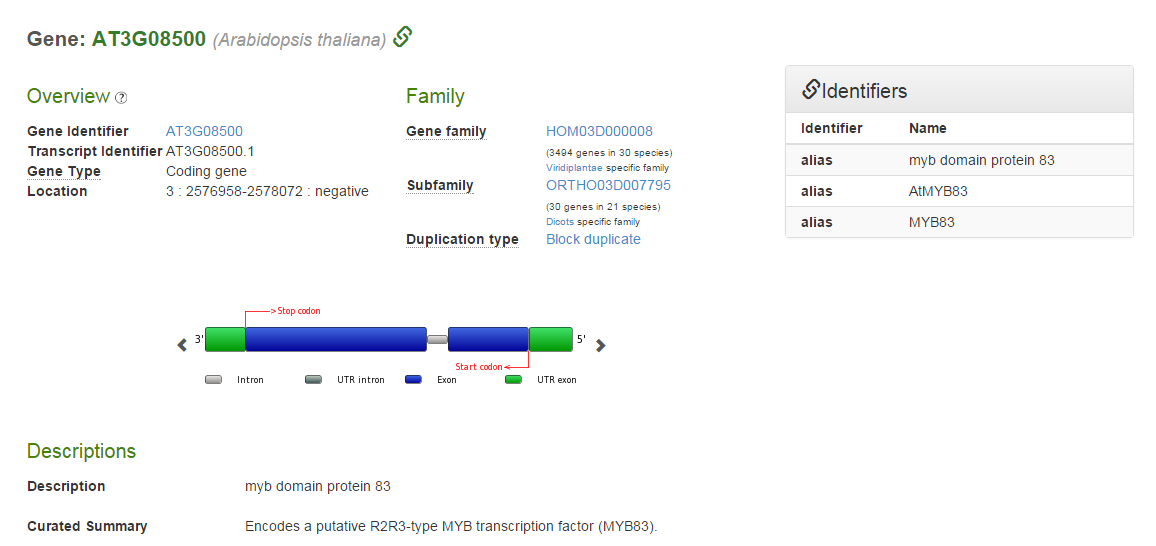
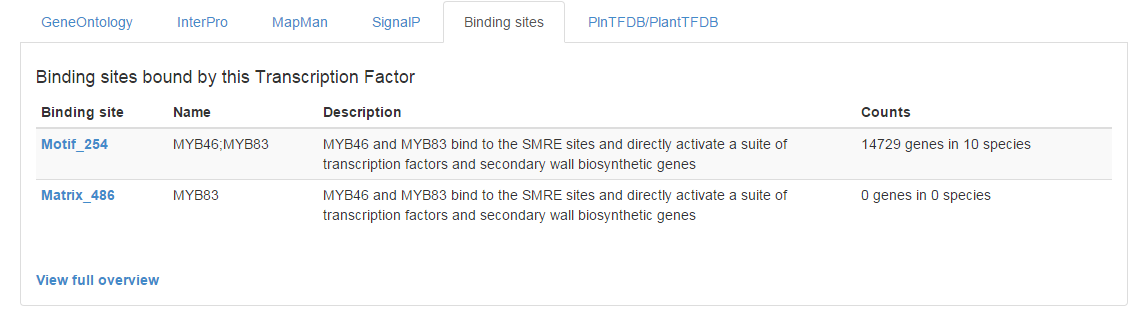
- For more detailed information on the target genes of a TF, the tab ‘Binding sites’ reports the associated binding site information. In this example MYB83 (AT3G08500), a TF involved in secondary cell wall related processes, is selected (see Figure 5).
- From this tab, a Binding site associated with the TF can be selected and information about the binding site as well as the predicted target genes is displayed. The Motif_254 is selected (see Figure 6).
- The targets for Arabidopsis thaliana can be selected from this page and downloaded into a separate text file (see Figure 7).
- This set of target genes can then be uploaded in the PLAZA workbench to perform GO enrichment on it (see workbench tutorial) (see Figure 8).
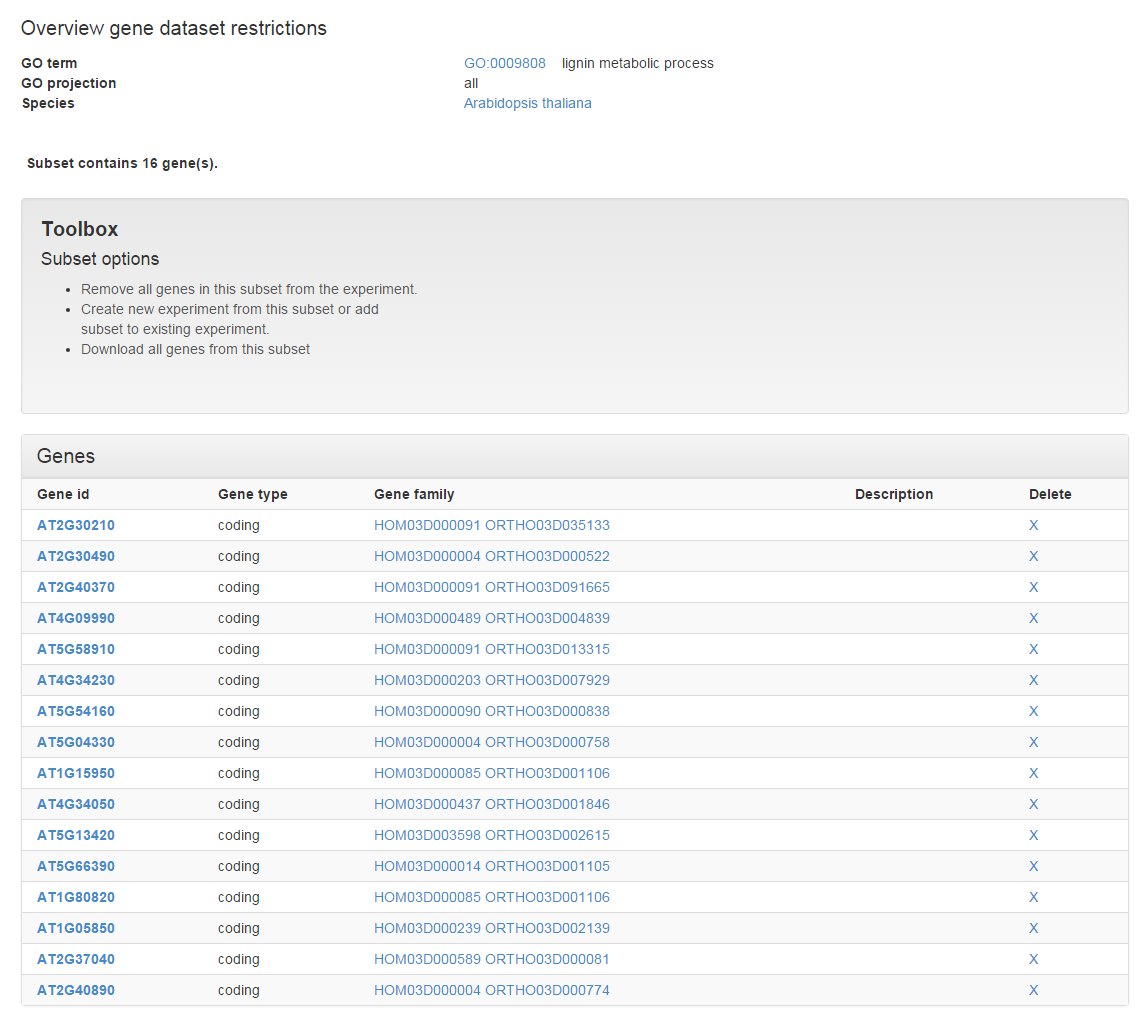
- From the list of enriched GO terms in the target gene set we can select the predicted target genes associated with a process of interest. In this case lignin metabolic process is selected, as lignin is an important component of the secondary cell wall (see Figure 9).